An engineered baculoviral protein and DNA co-delivery system for CRISPR-based mammalian genome editing
- PMID: 38412306
- PMCID: PMC11014373
- DOI: 10.1093/nar/gkae142
An engineered baculoviral protein and DNA co-delivery system for CRISPR-based mammalian genome editing
Abstract
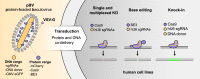
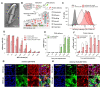
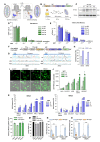
CRISPR-based DNA editing technologies enable rapid and accessible genome engineering of eukaryotic cells. However, the delivery of genetically encoded CRISPR components remains challenging and sustained Cas9 expression correlates with higher off-target activities, which can be reduced via Cas9-protein delivery. Here we demonstrate that baculovirus, alongside its DNA cargo, can be used to package and deliver proteins to human cells. Using protein-loaded baculovirus (pBV), we demonstrate delivery of Cas9 or base editors proteins, leading to efficient genome and base editing in human cells. By implementing a reversible, chemically inducible heterodimerization system, we show that protein cargoes can selectively and more efficiently be loaded into pBVs (spBVs). Using spBVs we achieved high levels of multiplexed genome editing in a panel of human cell lines. Importantly, spBVs maintain high editing efficiencies in absence of detectable off-targets events. Finally, by exploiting Cas9 protein and template DNA co-delivery, we demonstrate up to 5% site-specific targeted integration of a 1.8 kb heterologous DNA payload using a single spBV in a panel of human cell lines. In summary, we demonstrate that spBVs represent a versatile, efficient and potentially safer alternative for CRISPR applications requiring co-delivery of DNA and protein cargoes.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Baculoviral delivery of CRISPR/Cas9 facilitates efficient genome editing in human cells.PLoS One. 2017 Jun 22;12(6):e0179514. doi: 10.1371/journal.pone.0179514. eCollection 2017. PLoS One. 2017. PMID: 28640891 Free PMC article.
-
Highly efficient CRISPR-mediated large DNA docking and multiplexed prime editing using a single baculovirus.Nucleic Acids Res. 2022 Jul 22;50(13):7783-7799. doi: 10.1093/nar/gkac587. Nucleic Acids Res. 2022. PMID: 35801912 Free PMC article.
-
Delivery Aspects of CRISPR/Cas for in Vivo Genome Editing.Acc Chem Res. 2019 Jun 18;52(6):1555-1564. doi: 10.1021/acs.accounts.9b00106. Epub 2019 May 17. Acc Chem Res. 2019. PMID: 31099553 Free PMC article.
-
CRISPR/Cas9-based epigenome editing: An overview of dCas9-based tools with special emphasis on off-target activity.Methods. 2019 Jul 15;164-165:109-119. doi: 10.1016/j.ymeth.2019.05.003. Epub 2019 May 6. Methods. 2019. PMID: 31071448 Review.
-
Points of View on the Tools for Genome/Gene Editing.Int J Mol Sci. 2021 Sep 13;22(18):9872. doi: 10.3390/ijms22189872. Int J Mol Sci. 2021. PMID: 34576035 Free PMC article. Review.
Cited by
-
The Killer Saccharomyces cerevisiae Toxin: From Origin to Biomedical Research.Microorganisms. 2024 Dec 2;12(12):2481. doi: 10.3390/microorganisms12122481. Microorganisms. 2024. PMID: 39770684 Free PMC article. Review.
-
Enhancing Specificity, Precision, Accessibility, Flexibility, and Safety to Overcome Traditional CRISPR/Cas Editing Challenges and Shape Future Innovations.Adv Sci (Weinh). 2025 Jul;12(28):e2416331. doi: 10.1002/advs.202416331. Epub 2025 Jun 23. Adv Sci (Weinh). 2025. PMID: 40548648 Free PMC article. Review.
-
Tuning VSV-G Expression Improves Baculovirus Integrity, Stability and Mammalian Cell Transduction Efficiency.Viruses. 2024 Sep 17;16(9):1475. doi: 10.3390/v16091475. Viruses. 2024. PMID: 39339951 Free PMC article.
-
Structure of AcMNPV nucleocapsid reveals DNA portal organization and packaging apparatus of circular dsDNA baculovirus.Nat Commun. 2025 May 24;16(1):4844. doi: 10.1038/s41467-025-60152-2. Nat Commun. 2025. PMID: 40413174 Free PMC article.
-
Gene editing in hematopoietic stem cells by co-delivery of Cas9/sgRNA ribonucleoprotein and templates for homology-directed repair in 'all-in-one' lentivirus-derived nanoparticles.Nucleic Acids Res. 2025 Aug 11;53(15):gkaf767. doi: 10.1093/nar/gkaf767. Nucleic Acids Res. 2025. PMID: 40808298 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

