Evidence for dual targeting control of Arabidopsis 6-phosphogluconate dehydrogenase isoforms by N-terminal phosphorylation
- PMID: 38412416
- PMCID: PMC11103113
- DOI: 10.1093/jxb/erae077
Evidence for dual targeting control of Arabidopsis 6-phosphogluconate dehydrogenase isoforms by N-terminal phosphorylation
Abstract
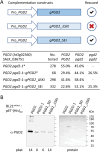
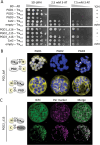
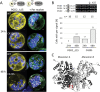
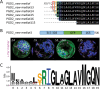
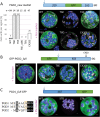
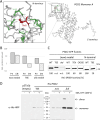
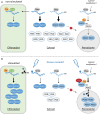
The oxidative pentose-phosphate pathway (OPPP) retrieves NADPH from glucose-6-phosphate, which is important in chloroplasts at night and in plastids of heterotrophic tissues. We previously studied how OPPP enzymes may transiently locate to peroxisomes, but how this is achieved for the third enzyme remained unclear. By extending our genetic approach, we demonstrated that Arabidopsis isoform 6-phosphogluconate dehydrogenase 2 (PGD2) is indispensable in peroxisomes during fertilization, and investigated why all PGD-reporter fusions show a mostly cytosolic pattern. A previously published interaction of a plant PGD with thioredoxin m was confirmed using Trxm2 for yeast two-hybrid (Y2H) and bimolecular fluorescent complementation (BiFC) assays, and medial reporter fusions (with both ends accessible) proved to be beneficial for studying peroxisomal targeting of PGD2. Of special importance were phosphomimetic changes at Thr6, resulting in a clear targeting switch to peroxisomes, while a similar change at position Ser7 in PGD1 conferred plastid import. Apparently, efficient subcellular localization can be achieved by activating an unknown kinase, either early after or during translation. N-terminal phosphorylation of PGD2 interfered with dimerization in the cytosol, thus allowing accessibility of the C-terminal peroxisomal targeting signal (PTS1). Notably, we identified amino acid positions that are conserved among plant PGD homologues, with PTS1 motifs first appearing in ferns, suggesting a functional link to fertilization during the evolution of seed plants.
Keywords: Arabidopsis PGD2; NADPH provision; OPPP; dual targeting; monomeric import; peroxisomes; protein phosphorylation.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures


Similar articles
-
Defects in Peroxisomal 6-Phosphogluconate Dehydrogenase Isoform PGD2 Prevent Gametophytic Interaction in Arabidopsis thaliana.Plant Physiol. 2016 May;171(1):192-205. doi: 10.1104/pp.15.01301. Epub 2016 Mar 3. Plant Physiol. 2016. PMID: 26941195 Free PMC article.
-
Analysis of homo- and hetero-dimerization among the three 6-phosphogluconate dehydrogenase isoforms of Arabidopsis.Plant Signal Behav. 2016 Oct 2;11(10):e1207034. doi: 10.1080/15592324.2016.1207034. Plant Signal Behav. 2016. PMID: 27366940 Free PMC article.
-
Alternative targeting of Arabidopsis plastidic glucose-6-phosphate dehydrogenase G6PD1 involves cysteine-dependent interaction with G6PD4 in the cytosol.Plant J. 2011 Jun;66(5):745-58. doi: 10.1111/j.1365-313X.2011.04535.x. Epub 2011 Mar 21. Plant J. 2011. PMID: 21309870
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
Cited by
-
Evidence for peroxisomal redundancy among the glucose-6-phosphate dehydrogenase isoforms of Arabidopsis thaliana.Plant Cell Physiol. 2025 May 30;66(5):722-737. doi: 10.1093/pcp/pcaf012. Plant Cell Physiol. 2025. PMID: 39829315 Free PMC article.
References
-
- Ackerman JD. 1993. Pollen germination and pollen tube growth in the marine angiosperm, Zostera marina L. Aquatic Botany 46, 189–202. doi:10.1016/0304-3770(93)90001-D - DOI
-
- Apanasets O, Grou CP, Van Veldhoven PP, Brees C, Wang B, Nordgren M, Dodt G, Azevedo JE, Fransen M.. 2014. PEX5, the shuttling import receptor for peroxisomal matrix proteins, is a redox-sensitive protein: PTS1 protein import and oxidative stress. Traffic 15, 94–103. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

