Biochemical and Structural Studies of the Carminomycin 4- O-Methyltransferase DnrK
- PMID: 38412432
- PMCID: PMC11623920
- DOI: 10.1021/acs.jnatprod.3c00947
Biochemical and Structural Studies of the Carminomycin 4- O-Methyltransferase DnrK
Abstract
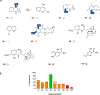
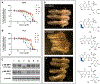
Structural and functional studies of the carminomycin 4-O-methyltransferase DnrK are described, with an emphasis on interrogating the acceptor substrate scope of DnrK. Specifically, the evaluation of 100 structurally and functionally diverse natural products and natural product mimetics revealed an array of pharmacophores as productive DnrK substrates. Representative newly identified DnrK substrates from this study included anthracyclines, angucyclines, anthraquinone-fused enediynes, flavonoids, pyranonaphthoquinones, and polyketides. The ligand-bound structure of DnrK bound to a non-native fluorescent hydroxycoumarin acceptor, 4-methylumbelliferone, along with corresponding DnrK kinetic parameters for 4-methylumbelliferone and native acceptor carminomycin are also reported for the first time. The demonstrated unique permissivity of DnrK highlights the potential for DnrK as a new tool in future biocatalytic and/or strain engineering applications. In addition, the comparative bioactivity assessment (cancer cell line cytotoxicity, 4E-BP1 phosphorylation, and axolotl embryo tail regeneration) of a select set of DnrK substrates/products highlights the ability of anthracycline 4-O-methylation to dictate diverse functional outcomes.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

