Tertiary Plasticity Drives the Efficiency of Enterocin 7B Interactions with Lipid Membranes
- PMID: 38412510
- PMCID: PMC10926100
- DOI: 10.1021/acs.jpcb.3c08199
Tertiary Plasticity Drives the Efficiency of Enterocin 7B Interactions with Lipid Membranes
Abstract
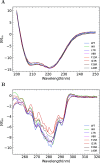
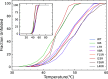
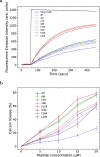
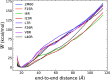
The ability of antimicrobial peptides to efficiently kill their bacterial targets depends on the efficiency of their binding to the microbial membrane. In the case of enterocins, there is a three-part interaction: initial binding, unpacking of helices on the membrane surface, and permeation of the lipid bilayer. Helical unpacking is driven by disruption of the peptide hydrophobic core when in contact with membranes. Enterocin 7B is a leaderless enterocin antimicrobial peptide produced from Enterococcus faecalis that functions alone, or with its cognate partner enterocin 7A, to efficiently kill a wide variety of Gram-stain positive bacteria. To better characterize the role that tertiary structural plasticity plays in the ability of enterocin 7B to interact with the membranes, a series of arginine single-site mutants were constructed that destabilize the hydrophobic core to varying degrees. A series of experimental measures of structure, stability, and function, including CD spectra, far UV CD melting profiles, minimal inhibitory concentrations analysis, and release kinetics of calcein, show that decreased stabilization of the hydrophobic core is correlated with increased efficiency of a peptide to permeate membranes and in killing bacteria. Finally, using the computational technique of adaptive steered molecular dynamics, we found that the atomistic/energetic landscape of peptide mechanical unfolding leads to free energy differences between the wild type and its mutants, whose trends correlate well with our experiment.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Solution structures of the linear leaderless bacteriocins enterocin 7A and 7B resemble carnocyclin A, a circular antimicrobial peptide.Biochemistry. 2013 Jun 11;52(23):3987-94. doi: 10.1021/bi400359z. Epub 2013 May 31. Biochemistry. 2013. PMID: 23725536
-
Production of enterocins L50A, L50B, and IT, a new enterocin, by Enterococcus faecium IT62, a strain isolated from Italian ryegrass in Japan.Antimicrob Agents Chemother. 2008 Jun;52(6):1917-23. doi: 10.1128/AAC.01409-07. Epub 2008 Apr 7. Antimicrob Agents Chemother. 2008. PMID: 18391036 Free PMC article.
-
Isolation and characterization of enterocin W, a novel two-peptide lantibiotic produced by Enterococcus faecalis NKR-4-1.Appl Environ Microbiol. 2012 Feb;78(3):900-3. doi: 10.1128/AEM.06497-11. Epub 2011 Dec 2. Appl Environ Microbiol. 2012. PMID: 22138996 Free PMC article.
-
Enterocins: Classification, Synthesis, Antibacterial Mechanisms and Food Applications.Molecules. 2022 Mar 30;27(7):2258. doi: 10.3390/molecules27072258. Molecules. 2022. PMID: 35408657 Free PMC article. Review.
-
The two-peptide class II bacteriocins: structure, production, and mode of action.J Mol Microbiol Biotechnol. 2007;13(4):210-9. doi: 10.1159/000104750. J Mol Microbiol Biotechnol. 2007. PMID: 17827971 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

