Hexasomal particles: consequence or also consequential?
- PMID: 38412564
- PMCID: PMC11893180
- DOI: 10.1016/j.gde.2024.102163
Hexasomal particles: consequence or also consequential?
Abstract
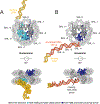
It is long known that an RNA polymerase transcribing through a nucleosome can generate subnucleosomal particles called hexasomes. These particles lack an H2A-H2B dimer, breaking the symmetry of a nucleosome and revealing new interfaces. Whether hexasomes are simply a consequence of RNA polymerase action or they also have a regulatory impact remains an open question. Recent biochemical and structural studies of RNA polymerases and chromatin remodelers with hexasomes motivated us to revisit this question. Here, we build on previous models to discuss how formation of hexasomes can allow sophisticated regulation of transcription and also significantly impact chromatin folding. We anticipate that further cellular and biochemical analysis of these subnucleosomal particles will uncover additional regulatory roles.
Copyright © 2024 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflict of interest.
Figures


References
-
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D: FACT facilitates transcription-dependent nucleosome alteration. Science 2003, 301:1090–1093. - PubMed
-
- Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM: Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell 2002, 9:541–552. - PubMed
-
- Baer BW, Rhodes D: Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature 1983, 301:482–488. - PubMed

