A comparative analysis of peptide-delivered antisense antibiotics using diverse nucleotide mimics
- PMID: 38413166
- PMCID: PMC11098465
- DOI: 10.1261/rna.079969.124
A comparative analysis of peptide-delivered antisense antibiotics using diverse nucleotide mimics
Abstract
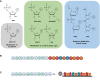
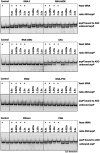
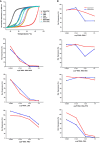
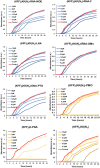
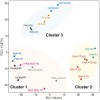
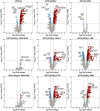
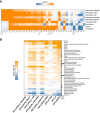
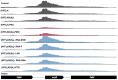
Antisense oligomer (ASO)-based antibiotics that target mRNAs of essential bacterial genes have great potential for counteracting antimicrobial resistance and for precision microbiome editing. To date, the development of such antisense antibiotics has primarily focused on using phosphorodiamidate morpholino (PMO) and peptide nucleic acid (PNA) backbones, largely ignoring the growing number of chemical modalities that have spurred the success of ASO-based human therapy. Here, we directly compare the activities of seven chemically distinct 10mer ASOs, all designed to target the essential gene acpP upon delivery with a KFF-peptide carrier into Salmonella. Our systematic analysis of PNA, PMO, phosphorothioate (PTO)-modified DNA, 2'-methylated RNA (RNA-OMe), 2'-methoxyethylated RNA (RNA-MOE), 2'-fluorinated RNA (RNA-F), and 2'-4'-locked RNA (LNA) is based on a variety of in vitro and in vivo methods to evaluate ASO uptake, target pairing and inhibition of bacterial growth. Our data show that only PNA and PMO are efficiently delivered by the KFF peptide into Salmonella to inhibit bacterial growth. Nevertheless, the strong target binding affinity and in vitro translational repression activity of LNA and RNA-MOE make them promising modalities for antisense antibiotics that will require the identification of an effective carrier.
Keywords: RNA-seq; antibiotics; antimicrobial resistance; antisense oligomers; nucleic acid mimics.
© 2024 Ghosh et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


References
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
-
- Bost JP, Barriga H, Holme MN, Gallud A, Maugeri M, Gupta D, Lehto T, Valadi H, Esbjörner EK, Stevens MM, et al. 2021. Delivery of oligonucleotide therapeutics: chemical modifications, lipid nanoparticles, and extracellular vesicles. ACS Nano 15: 13993–14021. 10.1021/acsnano.1c05099 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
