Convergent and divergent genes expression profiles associated with brain-wide functional connectome dysfunction in deficit and non-deficit schizophrenia
- PMID: 38413564
- PMCID: PMC10899251
- DOI: 10.1038/s41398-024-02827-w
Convergent and divergent genes expression profiles associated with brain-wide functional connectome dysfunction in deficit and non-deficit schizophrenia
Abstract
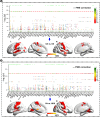
Deficit schizophrenia (DS) is a subtype of schizophrenia characterized by the primary and persistent negative symptoms. Previous studies have identified differences in brain functions between DS and non-deficit schizophrenia (NDS) patients. However, the genetic regulation features underlying these abnormal changes are still unknown. This study aimed to detect the altered patterns of functional connectivity (FC) in DS and NDS and investigate the gene expression profiles underlying these abnormal FC. The study recruited 82 DS patients, 96 NDS patients, and 124 healthy controls (CN). Voxel-based unbiased brain-wide association study was performed to reveal altered patterns of FC in DS and NDS patients. Machine learning techniques were used to access the utility of altered FC for diseases diagnosis. Weighted gene co-expression network analysis (WGCNA) was employed to explore the associations between altered FC and gene expression of 6 donated brains. Enrichment analysis was conducted to identify the genetic profiles, and the spatio-temporal expression patterns of the key genes were further explored. Comparing to CN, 23 and 20 brain regions with altered FC were identified in DS and NDS patients. The altered FC among these regions showed significant correlations with the SDS scores and exhibited high efficiency in disease classification. WGCNA revealed associations between DS/NDS-related gene expression and altered FC. Additionally, 22 overlapped genes, including 12 positive regulation genes and 10 negative regulation genes, were found between NDS and DS. Enrichment analyses demonstrated relationships between identified genes and significant pathways related to cellular response, neuro regulation, receptor binding, and channel activity. Spatial and temporal gene expression profiles of SCN1B showed the lowest expression at the initiation of embryonic development, while DPYSL3 exhibited rapid increased in the fetal. The present study revealed different altered patterns of FC in DS and NDS patients and highlighted the potential value of FC in disease classification. The associations between gene expression and neuroimaging provided insights into specific and common genetic regulation underlying these brain functional changes in DS and NDS, suggesting a potential genetic-imaging pathogenesis of schizophrenia.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Altered patterns of dynamic functional connectivity of brain networks in deficit and non-deficit schizophrenia.Eur Arch Psychiatry Clin Neurosci. 2025 Apr;275(3):743-753. doi: 10.1007/s00406-024-01803-1. Epub 2024 Apr 25. Eur Arch Psychiatry Clin Neurosci. 2025. PMID: 38662092
-
Convergent and divergent altered patterns of default mode network in deficit and non-deficit schizophrenia.Prog Neuropsychopharmacol Biol Psychiatry. 2019 Mar 8;89:427-434. doi: 10.1016/j.pnpbp.2018.10.012. Epub 2018 Oct 24. Prog Neuropsychopharmacol Biol Psychiatry. 2019. PMID: 30367960
-
Using support vector machine to explore the difference of function connection between deficit and non-deficit schizophrenia based on gray matter volume.Front Neurosci. 2023 Mar 27;17:1132607. doi: 10.3389/fnins.2023.1132607. eCollection 2023. Front Neurosci. 2023. PMID: 37051145 Free PMC article.
-
Functional Connectivity Biomarkers in Schizophrenia.Adv Neurobiol. 2024;40:237-283. doi: 10.1007/978-3-031-69491-2_10. Adv Neurobiol. 2024. PMID: 39562448 Review.
-
Cerebellum in neurodegenerative diseases: Advances, challenges, and prospects.iScience. 2024 Oct 18;27(11):111194. doi: 10.1016/j.isci.2024.111194. eCollection 2024 Nov 15. iScience. 2024. PMID: 39555407 Free PMC article. Review.
Cited by
-
Left amygdala alterations mediate the effects of negative symptoms on social dysfunction in schizophrenia.Schizophrenia (Heidelb). 2025 Jul 29;11(1):107. doi: 10.1038/s41537-025-00655-5. Schizophrenia (Heidelb). 2025. PMID: 40730820 Free PMC article.
-
LncRNA-miRNA‒mRNA Network in Schizophrenia.J Mol Neurosci. 2025 Aug 12;75(3):104. doi: 10.1007/s12031-025-02397-6. J Mol Neurosci. 2025. PMID: 40794250
-
Age-Specific Functional Connectivity Changes After Partial Sleep Deprivation Are Correlated With Neurocognitive and Molecular Signatures.CNS Neurosci Ther. 2025 Feb;31(2):e70272. doi: 10.1111/cns.70272. CNS Neurosci Ther. 2025. PMID: 39932149 Free PMC article.
-
Relationship between negative symptoms, cognitive function and social function in schizophrenia: new insight from a network analysis.Front Psychiatry. 2025 Jun 26;16:1623147. doi: 10.3389/fpsyt.2025.1623147. eCollection 2025. Front Psychiatry. 2025. PMID: 40642413 Free PMC article.
References
-
- Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, et al. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet. 2013;382:1575–86. - PubMed
-
- Tandon R, Nasrallah HA, Keshavan MS. Schizophrenia, “just the facts” 5. Treatment and prevention. Past, present, and future. Schizophr Res. 2010;122:1–23. - PubMed
-
- Mucci A, Merlotti E, Ucok A, Aleman A, Galderisi S. Primary and persistent negative symptoms: concepts, assessments and neurobiological bases. Schizophr Res. 2017;186:19–28. - PubMed
-
- Carpenter WT, Jr, Heinrichs DW, Wagman AM. Deficit and nondeficit forms of schizophrenia: the concept. Am J Psychiatry. 1988;145:578–83. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

