Inhibition of S6K lowers age-related inflammation and increases lifespan through the endolysosomal system
- PMID: 38413780
- PMCID: PMC11031405
- DOI: 10.1038/s43587-024-00578-3
Inhibition of S6K lowers age-related inflammation and increases lifespan through the endolysosomal system
Abstract
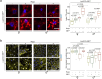
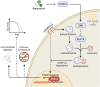
Suppression of target of rapamycin complex 1 (TORC1) by rapamycin ameliorates aging in diverse species. S6 kinase (S6K) is an essential mediator, but the mechanisms involved are unclear. Here we show that activation of S6K specifically in Drosophila fat-body blocked extension of lifespan by rapamycin, induced accumulation of multilamellar lysosomes and blocked age-associated hyperactivation of the NF-κB-like immune deficiency (IMD) pathway, indicative of reduced inflammaging. Syntaxin 13 mediated the effects of TORC1-S6K signaling on lysosome morphology and inflammaging, suggesting they may be linked. Inflammaging depended on the IMD receptor regulatory isoform PGRP-LC, and repression of the IMD pathway from midlife extended lifespan. Age-related inflammaging was higher in females than in males and was not lowered in males by rapamycin treatment or lowered S6K. Rapamycin treatment also elevated Syntaxin 12/13 levels in mouse liver and prevented age-related increase in noncanonical NF-κB signaling, suggesting that the effect of TORC1 on inflammaging is conserved from flies to mammals.
© 2024. Crown.
Conflict of interest statement
The authors declare no competing interests.
Figures











Similar articles
-
Rapamycin improves healthspan but not inflammaging in nfκb1-/- mice.Aging Cell. 2019 Feb;18(1):e12882. doi: 10.1111/acel.12882. Epub 2018 Nov 23. Aging Cell. 2019. PMID: 30468013 Free PMC article.
-
Neuronal TORC1 modulates longevity via AMPK and cell nonautonomous regulation of mitochondrial dynamics in C. elegans.Elife. 2019 Aug 14;8:e49158. doi: 10.7554/eLife.49158. Elife. 2019. PMID: 31411562 Free PMC article.
-
Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster.Cell Metab. 2010 Jan;11(1):35-46. doi: 10.1016/j.cmet.2009.11.010. Cell Metab. 2010. PMID: 20074526 Free PMC article.
-
TOR action in mammalian cells and in Caenorhabditis elegans.Curr Top Microbiol Immunol. 2004;279:115-38. doi: 10.1007/978-3-642-18930-2_8. Curr Top Microbiol Immunol. 2004. PMID: 14560955 Review.
-
Metabolic regulation, mitochondria and the life-prolonging effect of rapamycin: a mini-review.Gerontology. 2012;58(6):524-30. doi: 10.1159/000342204. Epub 2012 Aug 30. Gerontology. 2012. PMID: 22947849 Review.
Cited by
-
Inflammageing, a targetable pathway for preventing cardiovascular diseases.Cardiovasc Res. 2025 Aug 28;121(10):1537-1550. doi: 10.1093/cvr/cvae240. Cardiovasc Res. 2025. PMID: 39530590 Free PMC article. Review.
-
Ribosomal S6 kinase 1 regulates inflammaging via the senescence secretome.Nat Aging. 2024 Nov;4(11):1544-1561. doi: 10.1038/s43587-024-00695-z. Epub 2024 Aug 29. Nat Aging. 2024. PMID: 39210150 Free PMC article.
-
Functional characterisation of rare variants in genes encoding the MAPK/ERK signalling pathway identified in long-lived Leiden Longevity Study participants.Geroscience. 2025 May 29. doi: 10.1007/s11357-025-01699-2. Online ahead of print. Geroscience. 2025. PMID: 40442464
-
Gut aging: A wane from the normal to repercussion and gerotherapeutic strategies.Heliyon. 2024 Sep 12;10(19):e37883. doi: 10.1016/j.heliyon.2024.e37883. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39381110 Free PMC article. Review.
-
Rejuvenation Strategy for Inducing and Enhancing Autoimmune Response to Eliminate Senescent Cells.Aging Dis. 2024 Jul 17;16(4):2273-2292. doi: 10.14336/AD.2024.0579. Aging Dis. 2024. PMID: 39122450 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

