PMF-seq: a highly scalable screening strategy for linking genetics to mitochondrial bioenergetics
- PMID: 38413804
- PMCID: PMC11052718
- DOI: 10.1038/s42255-024-00994-0
PMF-seq: a highly scalable screening strategy for linking genetics to mitochondrial bioenergetics
Abstract
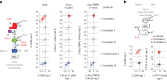
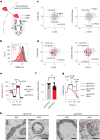
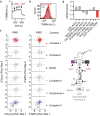
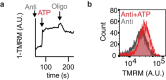
Our current understanding of mitochondrial organelle physiology has benefited from two broad approaches: classically, cuvette-based measurements with suspensions of isolated mitochondria, in which bioenergetic parameters are monitored acutely in response to respiratory chain substrates and inhibitors1-4, and more recently, highly scalable genetic screens for fitness phenotypes associated with coarse-grained properties of the mitochondrial state5-10. Here we introduce permeabilized-cell mitochondrial function sequencing (PMF-seq) to combine strengths of these two approaches to connect genes to detailed bioenergetic phenotypes. In PMF-seq, the plasma membranes within a pool of CRISPR mutagenized cells are gently permeabilized under conditions that preserve mitochondrial physiology, where detailed bioenergetics can be probed in the same way as with isolated organelles. Cells with desired bioenergetic parameters are selected optically using flow cytometry and subjected to next-generation sequencing. Using PMF-seq, we recover genes differentially required for mitochondrial respiratory chain branching and reversibility. We demonstrate that human D-lactate dehydrogenase specifically conveys electrons from D-lactate into cytochrome c to support mitochondrial membrane polarization. Finally, we screen for genetic modifiers of tBID, a pro-apoptotic protein that acts directly and acutely on mitochondria. We find the loss of the complex V assembly factor ATPAF2 acts as a genetic sensitizer of tBID's acute action. We anticipate that PMF-seq will be valuable for defining genes critical to the physiology of mitochondria and other organelles.
© 2024. The Author(s).
Conflict of interest statement
V.K.M. is a paid advisor to 5am Ventures. T.-L.T. and V.K.M. are listed as inventors on a provisional patent application filed by the Broad Institute on PMF-seq technology. The other authors declare no competing interests.
Figures




References
-
- Nicholls, D. G. & Ferguson, S. J. Bioenergetics 4th edn (Elsevier, 2013).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

