Histone butyrylation in the mouse intestine is mediated by the microbiota and associated with regulation of gene expression
- PMID: 38413806
- PMCID: PMC11520355
- DOI: 10.1038/s42255-024-00992-2
Histone butyrylation in the mouse intestine is mediated by the microbiota and associated with regulation of gene expression
Abstract
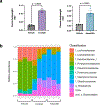
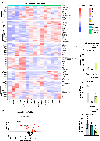
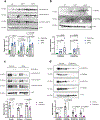
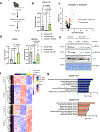
Post-translational modifications (PTMs) on histones are a key source of regulation on chromatin through impacting cellular processes, including gene expression1. These PTMs often arise from metabolites and are thus impacted by metabolism and environmental cues2-7. One class of metabolically regulated PTMs are histone acylations, which include histone acetylation, butyrylation, crotonylation and propionylation3,8. As these PTMs can be derived from short-chain fatty acids, which are generated by the commensal microbiota in the intestinal lumen9-11, we aimed to define how microbes impact the host intestinal chromatin landscape, mainly in female mice. Here we show that in addition to acetylation, intestinal epithelial cells from the caecum and distal mouse intestine also harbour high levels of butyrylation and propionylation on lysines 9 and 27 of histone H3. We demonstrate that these acylations are regulated by the microbiota and that histone butyrylation is additionally regulated by the metabolite tributyrin. Tributyrin-regulated gene programmes are correlated with histone butyrylation, which is associated with active gene-regulatory elements and levels of gene expression. Together, our study uncovers a regulatory layer of how the microbiota and metabolites influence the intestinal epithelium through chromatin, demonstrating a physiological setting in which histone acylations are dynamically regulated and associated with gene regulation.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests Statement
The authors declare no competing interests.
Figures








References
-
- Allis CD & Jenuwein T The molecular hallmarks of epigenetic control. Nat Rev Genet 17, 487–500 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RFA 598467/Crohn's and Colitis Foundation (Crohn's & Colitis Foundation)
- F32GM134560/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- K99GM143550/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- K99 GM143550/GM/NIGMS NIH HHS/United States
- R01 HD106051/HD/NICHD NIH HHS/United States
- R00 GM143550/GM/NIGMS NIH HHS/United States
- R01 AI118891/AI/NIAID NIH HHS/United States
- R01HD106051/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- R01AI118891/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- T32 CA009140/CA/NCI NIH HHS/United States
- F32 GM134560/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

