Characterizing complete mitochondrial genome of Aquilegia amurensis and its evolutionary implications
- PMID: 38413922
- PMCID: PMC10900605
- DOI: 10.1186/s12870-024-04844-9
Characterizing complete mitochondrial genome of Aquilegia amurensis and its evolutionary implications
Abstract
Background: Aquilegia is a model system for studying the evolution of adaptive radiation. However, very few studies have been conducted on the Aquilegia mitochondrial genome. Since mitochondria play a key role in plant adaptation to abiotic stress, analyzing the mitochondrial genome may provide a new perspective for understanding adaptive evolution.
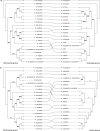
Results: The Aquilegia amurensis mitochondrial genome was characterized by a circular chromosome and two linear chromosomes, with a total length of 538,736 bp; the genes included 33 protein-coding genes, 24 transfer RNA (tRNA) genes and 3 ribosomal RNA (rRNA) genes. We subsequently conducted a phylogenetic analysis based on single nucleotide polymorphisms (SNPs) in the mitochondrial genomes of 18 Aquilegia species, which were roughly divided into two clades: the European-Asian clade and the North American clade. Moreover, the genes mttB and rpl5 were shown to be positively selected in European-Asian species, and they may help European and Asian species adapt to environmental changes.
Conclusions: In this study, we assembled and annotated the first mitochondrial genome of the adaptive evolution model plant Aquilegia. The subsequent analysis provided us with a basis for further molecular studies on Aquilegia mitochondrial genomes and valuable information on adaptive evolution in Aquilegia.
Keywords: Aquilegia; Mitochondrial genome; Phylogenetic analysis; Selection pressure analysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Comparative chloroplast genomes and phylogenetic analysis of Aquilegia.Appl Plant Sci. 2021 Mar 16;9(3):e11412. doi: 10.1002/aps3.11412. eCollection 2021 Mar. Appl Plant Sci. 2021. PMID: 33854846 Free PMC article.
-
The complete mitochondrial genome of Vanessa indica and phylogenetic analyses of the family Nymphalidae.Genes Genomics. 2018 Oct;40(10):1011-1022. doi: 10.1007/s13258-018-0709-x. Epub 2018 Jun 14. Genes Genomics. 2018. PMID: 29949077
-
Assembly and comparative analysis of the complete mitochondrial genome of Isopyrum anemonoides (Ranunculaceae).PLoS One. 2023 Oct 5;18(10):e0286628. doi: 10.1371/journal.pone.0286628. eCollection 2023. PLoS One. 2023. PMID: 37796878 Free PMC article.
-
Assembly and comparative analysis of the complete mitochondrial genome of white towel gourd (Luffa cylindrica).Genomics. 2024 May;116(3):110859. doi: 10.1016/j.ygeno.2024.110859. Epub 2024 May 13. Genomics. 2024. PMID: 38750703 Review.
-
Adaptive radiations: from field to genomic studies.Proc Natl Acad Sci U S A. 2009 Jun 16;106 Suppl 1(Suppl 1):9947-54. doi: 10.1073/pnas.0901594106. Epub 2009 Jun 15. Proc Natl Acad Sci U S A. 2009. PMID: 19528644 Free PMC article. Review.
Cited by
-
Comparative analysis of the complete mitochondrial genomes of Firmiana danxiaensis and F. kwangsiensis (Malvaceae), two endangered Firmiana species in China.Planta. 2025 Apr 9;261(5):107. doi: 10.1007/s00425-025-04685-2. Planta. 2025. PMID: 40205193
-
Assembly and comparative analysis of the complete mitochondrial genome of Brassica rapa var. Purpuraria.BMC Genomics. 2024 Jun 1;25(1):546. doi: 10.1186/s12864-024-10457-1. BMC Genomics. 2024. PMID: 38824587 Free PMC article.
-
Mitochondrial genome analysis of the endangered Oreocharis esquirolii: insights into evolutionary adaptation and conservation.BMC Plant Biol. 2025 Jul 2;25(1):827. doi: 10.1186/s12870-025-06838-7. BMC Plant Biol. 2025. PMID: 40604449 Free PMC article.
-
Complete mitochondrial genome of Hippophae tibetana: insights into adaptation to high-altitude environments.Front Plant Sci. 2024 Aug 7;15:1449606. doi: 10.3389/fpls.2024.1449606. eCollection 2024. Front Plant Sci. 2024. PMID: 39170791 Free PMC article.
-
Comparative analysis of mitochondrial and chloroplast genomes of Dracaena cambodiana from contrasting island habitats.Front Plant Sci. 2025 Jun 25;16:1620721. doi: 10.3389/fpls.2025.1620721. eCollection 2025. Front Plant Sci. 2025. PMID: 40636010 Free PMC article.
References
-
- Jang W, Lee HO, Kim J-U, Lee J-W, Hong C-E, Bang K-H, Chung J-W, Jo I-HJA. Complete mitochondrial genome and a set of 10 novel kompetitive allele-specific PCR markers in ginseng (Panax Ginseng CA Mey.). 2020, 10(12):1868.
-
- Varré J-S, d’Agostino N, Touzet P, Gallina S, Tamburino R, Cantarella C, Ubrig E, Cardi T, Drouard L, Gualberto JM. Complete sequence, multichromosomal architecture and transcriptome analysis of the Solanum tuberosum mitochondrial genome. Int J Mol Sci. 2019;20(19):4788. doi: 10.3390/ijms20194788. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

