Splicing targeting drugs highlight intron retention as an actionable vulnerability in advanced prostate cancer
- PMID: 38413979
- PMCID: PMC10898177
- DOI: 10.1186/s13046-024-02986-0
Splicing targeting drugs highlight intron retention as an actionable vulnerability in advanced prostate cancer
Abstract
Background: Advanced prostate cancer (PC) is characterized by insensitivity to androgen deprivation therapy and chemotherapy, resulting in poor outcome for most patients. Thus, advanced PC urgently needs novel therapeutic strategies. Mounting evidence points to splicing dysregulation as a hallmark of advanced PC. Moreover, pharmacologic inhibition of the splicing process is emerging as a promising option for this disease.
Method: By using a representative androgen-insensitive PC cell line (22Rv1), we have investigated the genome-wide transcriptomic effects underlying the cytotoxic effects exerted by three splicing-targeting drugs: Pladienolide B, indisulam and THZ531. Bioinformatic analyses were performed to uncover the gene structural features underlying sensitivity to transcriptional and splicing regulation by these treatments. Biological pathways altered by these treatments were annotated by gene ontology analyses and validated by functional experiments in cell models.
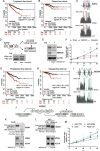
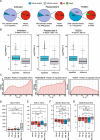
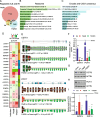
Results: Although eliciting similar cytotoxic effects on advanced PC cells, Pladienolide B, indisulam and THZ531 modulate specific transcriptional and splicing signatures. Drug sensitivity is associated with distinct gene structural features, expression levels and cis-acting sequence elements in the regulated exons and introns. Importantly, we identified PC-relevant genes (i.e. EZH2, MDM4) whose drug-induced splicing alteration exerts an impact on cell survival. Moreover, computational analyses uncovered a widespread impact of splicing-targeting drugs on intron retention, with enrichment in genes implicated in pre-mRNA 3'-end processing (i.e. CSTF3, PCF11). Coherently, advanced PC cells displayed high sensitivity to a specific inhibitor of the cleavage and polyadenylation complex, which enhances the effects of chemotherapeutic drugs that are already in use for this cancer.
Conclusions: Our study uncovers intron retention as an actionable vulnerability for advanced PC, which may be exploited to improve therapeutic management of this currently incurable disease.
Keywords: 3’-end mRNA processing; Advanced prostate cancer; Alternative splicing; Intron-retention; Splicing inhibitors; Transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
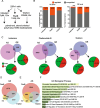
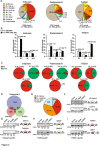
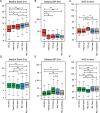
Our study highlights the cis-acting regulatory features underlying susceptibility to transcriptional and post-transcriptional variations induced by splicing-targeting drugs; it reveals IR as the prominent pattern induced by both direct (Ind/PlaB) and indirect (THZ531) inhibition of splicing and uncovers the 3’-end mRNA processing as a druggable vulnerability for advanced PC.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

