Targeting of mitochondrial fission through natural flavanones elicits anti-myeloma activity
- PMID: 38413989
- PMCID: PMC10898065
- DOI: 10.1186/s12967-024-05013-0
Targeting of mitochondrial fission through natural flavanones elicits anti-myeloma activity
Abstract
Background: Mitochondrial alterations, often dependent on unbalanced mitochondrial dynamics, feature in the pathobiology of human cancers, including multiple myeloma (MM). Flavanones are natural flavonoids endowed with mitochondrial targeting activities. Herein, we investigated the capability of Hesperetin (Hes) and Naringenin (Nar), two aglycones of Hesperidin and Naringin flavanone glycosides, to selectively target Drp1, a pivotal regulator of mitochondrial dynamics, prompting anti-MM activity.
Methods: Molecular docking analyses were performed on the crystallographic structure of Dynamin-1-like protein (Drp1), using Hes and Nar molecular structures. Cell viability and apoptosis were assessed in MM cell lines, or in co-culture systems with primary bone marrow stromal cells, using Cell Titer Glo and Annexin V-7AAD staining, respectively; clonogenicity was determined using methylcellulose colony assays. Transcriptomic analyses were carried out using the Ion AmpliSeq™ platform; mRNA and protein expression levels were determined by quantitative RT-PCR and western blotting, respectively. Mitochondrial architecture was assessed by transmission electron microscopy. Real time measurement of oxygen consumption was performed by high resolution respirometry in living cells. In vivo anti-tumor activity was evaluated in NOD-SCID mice subcutaneously engrafted with MM cells.
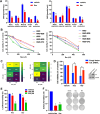
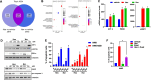
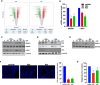
Results: Hes and Nar were found to accommodate within the GTPase binding site of Drp1, and to inhibit Drp1 expression and activity, leading to hyperfused mitochondria with reduced OXPHOS. In vitro, Hes and Nar reduced MM clonogenicity and viability, even in the presence of patient-derived bone marrow stromal cells, triggering ER stress and apoptosis. Interestingly, Hes and Nar rewired MM cell metabolism through the down-regulation of master transcriptional activators (SREBF-1, c-MYC) of lipogenesis genes. An extract of Tacle, a Citrus variety rich in Hesperidin and Naringin, was capable to recapitulate the phenotypic and molecular perturbations of each flavanone, triggering anti-MM activity in vivo.
Conclusion: Hes and Nar inhibit proliferation, rewire the metabolism and induce apoptosis of MM cells via antagonism of the mitochondrial fission driver Drp1. These results provide a framework for the development of natural anti-MM therapeutics targeting aberrant mitochondrial dependencies.
Keywords: Flavanones; Hesperitin; Mitochondrial dynamics; Multiple myeloma; Naringenin.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Kumar SK, Rajkumar V, Kyle RA, van Duin M, Sonneveld P, Mateos M-V, et al. Multiple myeloma. Nat Rev Dis Primers. 2017;3:17046. - PubMed
-
- Alsayyah C, Ozturk O, Cavellini L, Belgareh-Touzé N, Cohen MM. The regulation of mitochondrial homeostasis by the ubiquitin proteasome system. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2020;1861:148302. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

