Reduced representative methylome profiling of cell-free DNA for breast cancer detection
- PMID: 38414041
- PMCID: PMC10898043
- DOI: 10.1186/s13148-024-01641-x
Reduced representative methylome profiling of cell-free DNA for breast cancer detection
Abstract
Background: Whole-genome methylation sequencing of cfDNA is not cost-effective for tumor detection. Here, we introduce reduced representative methylome profiling (RRMP), which employs restriction enzyme for depletion of AT-rich sequence to achieve enrichment and deep sequencing of CG-rich sequences.
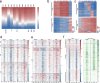
Methods: We first verified the ability of RRMP to enrich CG-rich sequences using tumor cell genomic DNA and analyzed differential methylation regions between tumor cells and normal whole blood cells. We then analyzed cfDNA from 29 breast cancer patients and 27 non-breast cancer individuals to detect breast cancer by building machine learning models.
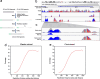
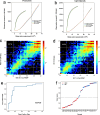
Results: RRMP captured 81.9% CpG islands and 75.2% gene promoters when sequenced to 10 billion base pairs, with an enrichment efficiency being comparable to RRBS. RRMP allowed us to assess DNA methylation changes between tumor cells and whole blood cells. Applying our approach to cfDNA from 29 breast cancer patients and 27 non-breast cancer individuals, we developed machine learning models that could discriminate between breast cancer and non-breast cancer controls (AUC = 0.85), suggesting possibilities for truly non-invasive cancer detection.
Conclusions: We developed a new method to achieve reduced representative methylome profiling of cell-free DNA for tumor detection.
© 2024. The Author(s).
Conflict of interest statement
Chen is the founder of Vangenes, Inc., X. Zhu, Y. Liu, Z. He, H. Xu, H. Zheng, Z. Huang, D. Wang, Z. Huang and X. Lin are employees of Vangenes, Inc. P. Guo is an employee of Huazao Biotechnology CO., LTD. The authors have filed a patent application on methods described in this manuscript (CN 113943779 A). The remaining authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

