Using a targeted metabolomics approach to explore differences in ARDS associated with COVID-19 compared to ARDS caused by H1N1 influenza and bacterial pneumonia
- PMID: 38414082
- PMCID: PMC10900651
- DOI: 10.1186/s13054-024-04843-0
Using a targeted metabolomics approach to explore differences in ARDS associated with COVID-19 compared to ARDS caused by H1N1 influenza and bacterial pneumonia
Abstract
Rationale: Acute respiratory distress syndrome (ARDS) is a life-threatening critical care syndrome commonly associated with infections such as COVID-19, influenza, and bacterial pneumonia. Ongoing research aims to improve our understanding of ARDS, including its molecular mechanisms, individualized treatment options, and potential interventions to reduce inflammation and promote lung repair.
Objective: To map and compare metabolic phenotypes of different infectious causes of ARDS to better understand the metabolic pathways involved in the underlying pathogenesis.
Methods: We analyzed metabolic phenotypes of 3 ARDS cohorts caused by COVID-19, H1N1 influenza, and bacterial pneumonia compared to non-ARDS COVID-19-infected patients and ICU-ventilated controls. Targeted metabolomics was performed on plasma samples from a total of 150 patients using quantitative LC-MS/MS and DI-MS/MS analytical platforms.
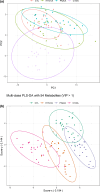
Results: Distinct metabolic phenotypes were detected between different infectious causes of ARDS. There were metabolomics differences between ARDSs associated with COVID-19 and H1N1, which include metabolic pathways involving taurine and hypotaurine, pyruvate, TCA cycle metabolites, lysine, and glycerophospholipids. ARDSs associated with bacterial pneumonia and COVID-19 differed in the metabolism of D-glutamine and D-glutamate, arginine, proline, histidine, and pyruvate. The metabolic profile of COVID-19 ARDS (C19/A) patients admitted to the ICU differed from COVID-19 pneumonia (C19/P) patients who were not admitted to the ICU in metabolisms of phenylalanine, tryptophan, lysine, and tyrosine. Metabolomics analysis revealed significant differences between C19/A, H1N1/A, and PNA/A vs ICU-ventilated controls, reflecting potentially different disease mechanisms.
Conclusion: Different metabolic phenotypes characterize ARDS associated with different viral and bacterial infections.
Keywords: Acute respiratory distress syndrome; H1N1; Metabolomics; Pneumonia; SARS-CoV-2.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest, financial or otherwise, except Dr. James Russell who discloses the following: COI: DR. JAMES A. RUSSELL AB, MD, FRCPC FINANCIAL DISCLOSURE (last 36 months). Dr. Russell reports patents owned by the University of British Columbia (UBC) that are related to (1) the use of PCSK9 inhibitor(s) in sepsis, (2) the use of vasopressin in septic shock, and (3) a patent owned by Ferring for the use of selepressin in septic shock. Dr. Russell is an inventor on these patents. Dr. Russell was a founder, Director, and shareholder in Cyon Therapeutics Inc. (now closed) and is a shareholder in Molecular You Corp. Dr. Russell is a Senior Research Advisor of the British Columbia, Canada Post COVID—Interdisciplinary Clinical Care Network (PC-ICCN). Dr. Russell is no longer actively consulting for any industry. Dr. Russell reports receiving consulting fees in the last three years from: (1) SIB Therapeutics LLC (developing a sepsis drug). (2) Ferring Pharmaceuticals (manufactures vasopressin and developing selepressin). (3) Dr. Russell was a funded member of the Data and Safety Monitoring Board (DSMB) of an NIH-sponsored plasma trial in COVID-19 (PASS-IT-ON) (2020–2021). (4) PAR Pharma (sells prepared bags of vasopressin). Dr. Russell reports receiving an investigator-initiated grant from Grifols (entitled "Is HBP a mechanism of albumin's efficacy in human septic shock?") that was provided to and administered by UBC. Dr. Russell has received grants for COVID-19 research: 4 from the Canadian Institutes of Health Research (CIHR) and three from the St. Paul's Foundation (SPF). Dr. Russell was a non-funded Science Advisor and member of the Government of Canada COVID-19 Therapeutics Task Force (June 2020–2021).
Figures


References
-
- Turner AJ. ACE2 Cell Biology, Regulation, and Physiological Functions. The Protective Arm of the Renin Angiotensin System (RAS). 2015:185–9.
-
- Burns KCMLTMASDTKLTMSBJSJWKPDLFMJHGF. Sustained Dysregulation of the Plasma Renin-angiotensin System in Acute COVID-19. 2021.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

