Real-time quantitative reverse transcription polymerase chain reaction detection of SARS-CoV-2 Delta variant in Canadian wastewater
- PMID: 38414535
- PMCID: PMC10898651
- DOI: 10.14745/ccdr.v49i05a07
Real-time quantitative reverse transcription polymerase chain reaction detection of SARS-CoV-2 Delta variant in Canadian wastewater
Abstract
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern are associated with increased infectivity, severity, and mortality of coronavirus disease 2019 (COVID-19) and have been increasingly detected in clinical and wastewater surveillance in Canada and internationally. In this study, we present a real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) assay for detection of the N gene D377Y mutation associated with the SARS-CoV-2 Delta variant in wastewater.
Methods: Wastewater samples (n=980) were collected from six cities and 17 rural communities across Canada from July to November 2021 and screened for the D377Y mutation.
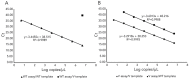
Results: The Delta variant was detected in all major Canadian cities and northern remote regions, and half of the southern rural communities. The sensitivity and specificity of this assay were sufficient for detection and quantitation of the Delta variant in wastewater to aid in rapid population-level screening and surveillance.
Conclusion: This study demonstrates a novel cost-effective RT-qPCR assay for tracking the spread of the SARS-CoV-2 Delta variant. This rapid assay can be easily integrated into current wastewater surveillance programs to aid in population-level variant tracking.
Keywords: B.1.617.2; Delta; SARS-CoV-2; qPCR; variant; wastewater.
Conflict of interest statement
Competing interests None.
Figures


References
-
- Naveca F, Nascimento V, Souza V, Corado A, Nascimento F, Silva G, Costa A, Duarte D, Pessoa K, Goncalves L, Brandao MJ, Jesus M, Fernandes C, Pinto R, Silva M, Mattos T, Wallau GL, Siqueira MM, Resende PC, Delatorre E, Graf T. Phylogenetic relationship of SARS-CoV-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the Spike protein. https://virological.org/t/phylogenetic-relationship-of-sars-cov-2-sequen...
-
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao NY, Korsman S, Davies MA, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LC, Kosakovsky Pond SL, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021;592(7854):438–43. 10.1038/s41586-021-03402-9 - DOI - PubMed
-
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Dabrera G, O’Toole Á, Amato R, Ragonnet-Cronin M, Harrison I, Jackson B, Ariani CV, Boyd O, Loman NJ, McCrone JT, Gonçalves S, Jorgensen D, Myers R, Hill V, Jackson DK, Gaythorpe K, Groves N, Sillitoe J, Kwiatkowski DP, Flaxman S, Ratmann O, Bhatt S, Hopkins S, Gandy A, Rambaut A, Ferguson NM; COVID-19 Genomics UK (COG-UK) consortium . Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 2021;593(7858):266–9. 10.1038/s41586-021-03470-x - DOI - PubMed
-
- World Health Organization Tracking SARS-CoV-2 Variants. Geneva, CH: WHO; 2023. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
LinkOut - more resources
Full Text Sources
Miscellaneous