Bioinformatics Analysis and Experimental Validation of Mitochondrial Autophagy Genes in Knee Osteoarthritis
- PMID: 38414629
- PMCID: PMC10898481
- DOI: 10.2147/IJGM.S444847
Bioinformatics Analysis and Experimental Validation of Mitochondrial Autophagy Genes in Knee Osteoarthritis
Abstract
Background: Mitochondrial autophagy is closely related to the pathogenesis of osteoarthritis, In order to explore the role of mitochondrial autophagy related genes in knee osteoarthritis (KOA) and its molecular mechanism.
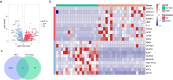
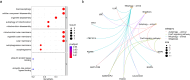
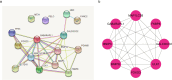
Methods: KOA-related transcriptome data were extracted from the Gene Expression Omnibus (GEO) database. Differentially expressed mitochondrial autophagy gene (DEMGs) were screened in patients with KOA by differential expression analysis. The STRING website was used to construct a protein-protein interaction (PPI) network among DEMGs. Molecular complex detection (MCODE) method in Cytoscape software was performed to identify hub DEMGs. Support vector machine recursive feature elimination (SVM-RFE) method was used to construct the hub DEMG diagnosis model. Genes with diagnostic value were identified as biomarkers by plotting receiver operating characteristic (ROC) curves and Expression validation. CIBERSORT algorithm was used to calculate the proportion of 22 immune cells in each sample in the GSE114007 dataset. Finally, biomarker expression was verified by qPCR.
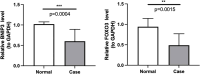
Results: A total of 15 DEMGs were obtained and enrichment analyses showed that these DEMG strains were mainly enriched in the mitophagy-animal, shigellosis, autophagy-animal and FoxO signal pathways. The PPI network unveiled 13 DEMGs with interactions. In addition, 8 hub DEMGs (ULK1, CALCOCO2, MAP1LC3B, BNIP3L, GABARAPL1, BNIP3, FKBP8 and FOXO3) were obtained for KOA. And 5 model DEMGs (BNIP3L, BNIP3, MAP1LC3B, ULK1 and FOXO3) were screened. The ROC curves revealed that BNIP3 and FOXO3 has strong diagnostic value in these models of DEMG. Immune-infiltration and correlation analysis showed that BNIP3 and FOXO3 were significantly correlated with three different immune cells, including primary B cells, M0 macrophage and M2 macrophage. The cartilage tissue samples qPCR verification results show that FOXO3 and BNIP3 were all down-regulated in KOA (p < 0.01), and the validation results are consistent with the above analysis.
Conclusion: BNIP3 and FOXO3 have been identified as biomarkers for the diagnosis of KOA, which might supply a new insight for the pathogenesis and treatment of KOA.
Keywords: Gene Expression Omnibus; bioinformatics analysis; biomarkers; diagnostic; immune infiltration.
© 2024 Tang et al.
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose for this work.
Figures


Similar articles
-
Identification of autophagy-related genes in osteoarthritis articular cartilage and their roles in immune infiltration.Front Immunol. 2023 Nov 27;14:1263988. doi: 10.3389/fimmu.2023.1263988. eCollection 2023. Front Immunol. 2023. PMID: 38090564 Free PMC article.
-
Identification of mitophagy-related biomarkers in human osteoporosis based on a machine learning model.Front Physiol. 2024 Jan 8;14:1289976. doi: 10.3389/fphys.2023.1289976. eCollection 2023. Front Physiol. 2024. PMID: 38260098 Free PMC article.
-
Renal tubular gen e biomarkers identification based on immune infiltrates in focal segmental glomerulosclerosis.Ren Fail. 2022 Dec;44(1):966-986. doi: 10.1080/0886022X.2022.2081579. Ren Fail. 2022. PMID: 35713363 Free PMC article.
-
Identification of novel mitophagy-related biomarkers for Kawasaki disease by integrated bioinformatics and machine-learning algorithms.Transl Pediatr. 2024 Aug 31;13(8):1439-1456. doi: 10.21037/tp-24-230. Epub 2024 Aug 26. Transl Pediatr. 2024. PMID: 39263286 Free PMC article.
-
Roles of the crucial mitochondrial DNA in hypertrophic cardiomyopathy prognosis and diagnosis: A review.Medicine (Baltimore). 2023 Dec 1;102(48):e36368. doi: 10.1097/MD.0000000000036368. Medicine (Baltimore). 2023. PMID: 38050313 Free PMC article. Review.
References
-
- Almonte-Becerril M, Navarro-Garcia F, Gonzalez-Robles A, Vega-Lopez MA, Lavalle C, Kouri JB. Cell death of chondrocytes is a combination between apoptosis and autophagy during the pathogenesis of Osteoarthritis within an experimental model. Apoptosis. 2010;15(5):631–638. doi:10.1007/s10495-010-0458-z - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

