Elucidating shared biomarkers in gastroesophageal reflux disease and idiopathic pulmonary fibrosis: insights into novel therapeutic targets and the role of angelicae sinensis radix
- PMID: 38414734
- PMCID: PMC10897002
- DOI: 10.3389/fphar.2024.1348708
Elucidating shared biomarkers in gastroesophageal reflux disease and idiopathic pulmonary fibrosis: insights into novel therapeutic targets and the role of angelicae sinensis radix
Abstract
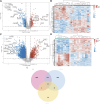
Background: The etiological underpinnings of gastroesophageal reflux disease (GERD) and idiopathic pulmonary fibrosis (IPF) remain elusive, coupled with a scarcity of effective therapeutic interventions for IPF. Angelicae sinensis radix (ASR, also named Danggui) is a Chinese herb with potential anti-fibrotic properties, that holds promise as a therapeutic agent for IPF. Objective: This study seeks to elucidate the causal interplay and potential mechanisms underlying the coexistence of GERD and IPF. Furthermore, it aims to investigate the regulatory effect of ASR on this complex relationship. Methods: A two-sample Mendelian randomization (TSMR) approach was employed to delineate the causal connection between gastroesophageal reflux disease and IPF, with Phennoscanner V2 employed to mitigate confounding factors. Utilizing single nucleotide polymorphism (SNPs) and publicly available microarray data, we analyzed potential targets and mechanisms related to IPF in GERD. Network pharmacology and molecular docking were employed to explore the targets and efficacy of ASR in treating GERD-related IPF. External datasets were subsequently utilized to identify potential diagnostic biomarkers for GERD-related IPF. Results: The IVW analysis demonstrated a positive causal relationship between GERD and IPF (IVW: OR = 1.002, 95%CI: 1.001, 1.003; p < 0.001). Twenty-five shared differentially expressed genes (DEGs) were identified. GO functional analysis revealed enrichment in neural, cellular, and brain development processes, concentrated in chromosomes and plasma membranes, with protein binding and activation involvement. KEGG analysis unveiled enrichment in proteoglycan, ERBB, and neuroactive ligand-receptor interaction pathways in cancer. Protein-protein interaction (PPI) analysis identified seven hub genes. Network pharmacology analysis demonstrated that 104 components of ASR targeted five hub genes (PDE4B, DRD2, ERBB4, ESR1, GRM8), with molecular docking confirming their excellent binding efficiency. GRM8 and ESR1 emerged as potential diagnostic biomarkers for GERD-related IPF (ESR1: AUCGERD = 0.762, AUCIPF = 0.725; GRM8: AUCGERD = 0.717, AUCIPF = 0.908). GRM8 and ESR1 emerged as potential diagnostic biomarkers for GERD-related IPF, validated in external datasets. Conclusion: This study establishes a causal link between GERD and IPF, identifying five key targets and two potential diagnostic biomarkers for GERD-related IPF. ASR exhibits intervention efficacy and favorable binding characteristics, positioning it as a promising candidate for treating GERD-related IPF. The potential regulatory mechanisms may involve cell responses to fibroblast growth factor stimulation and steroidal hormone-mediated signaling pathways.
Keywords: angelicae sinensis radix; gastroesophageal reflux disease; idiopathic pulmonary fibrosis; mendelian randomization; network-pharmacology.
Copyright © 2024 Wu, Xiao, Fang, He, Wang, Wang, Lan, Wang, Du and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

