Myoglobin modulates the Hippo pathway to promote cardiomyocyte differentiation
- PMID: 38414852
- PMCID: PMC10897895
- DOI: 10.1016/j.isci.2024.109146
Myoglobin modulates the Hippo pathway to promote cardiomyocyte differentiation
Abstract
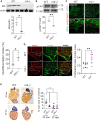
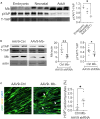
The endogenous mechanisms that propagate cardiomyocyte differentiation and prevent de-differentiation remain unclear. While the expression of the heme protein myoglobin increases by over 50% during cardiomyocyte differentiation, a role for myoglobin in regulating cardiomyocyte differentiation has not been tested. Here, we show that deletion of myoglobin in cardiomyocyte models decreases the gene expression of differentiation markers and stimulates cellular proliferation, consistent with cardiomyocyte de-differentiation. Mechanistically, the heme prosthetic group of myoglobin catalyzes the oxidation of the Hippo pathway kinase LATS1, resulting in phosphorylation and inactivation of yes-associated protein (YAP). In vivo, myoglobin-deficient zebrafish hearts show YAP dephosphorylation and accelerated cardiac regeneration after apical injury. Similarly, myoglobin knockdown in neonatal murine hearts shows increased YAP dephosphorylation and cardiomyocyte cycling. These data demonstrate a novel role for myoglobin as an endogenous driver of cardiomyocyte differentiation and highlight myoglobin as a potential target to enhance cardiac development and improve cardiac repair and regeneration.
Keywords: Biochemistry; Biological sciences; Cell biology; Natural sciences; Specialized functions of cells.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
gp130 Controls Cardiomyocyte Proliferation and Heart Regeneration.Circulation. 2020 Sep 8;142(10):967-982. doi: 10.1161/CIRCULATIONAHA.119.044484. Epub 2020 Jun 30. Circulation. 2020. PMID: 32600062
-
Calmodulin activates the Hippo signaling pathway by promoting LATS1 kinase-mediated inhibitory phosphorylation of the transcriptional coactivator YAP.J Biol Chem. 2022 May;298(5):101839. doi: 10.1016/j.jbc.2022.101839. Epub 2022 Mar 17. J Biol Chem. 2022. PMID: 35307353 Free PMC article.
-
Hippo Deficiency Leads to Cardiac Dysfunction Accompanied by Cardiomyocyte Dedifferentiation During Pressure Overload.Circ Res. 2019 Jan 18;124(2):292-305. doi: 10.1161/CIRCRESAHA.118.314048. Circ Res. 2019. PMID: 30582455 Free PMC article.
-
An emerging role for Hippo-YAP signaling in cardiovascular development.J Biomed Res. 2014 Jul;28(4):251-4. doi: 10.7555/JBR.28.20140020. Epub 2014 Mar 22. J Biomed Res. 2014. PMID: 25050107 Free PMC article. Review.
-
Upstream regulation of the Hippo-Yap pathway in cardiomyocyte regeneration.Semin Cell Dev Biol. 2020 Apr;100:11-19. doi: 10.1016/j.semcdb.2019.09.004. Epub 2019 Oct 9. Semin Cell Dev Biol. 2020. PMID: 31606277 Free PMC article. Review.
Cited by
-
Cannabidiol Ameliorates Doxorubicin-Induced Myocardial Injury via Activating Hippo Pathway.Drug Des Devel Ther. 2025 Jan 24;19:569-583. doi: 10.2147/DDDT.S497323. eCollection 2025. Drug Des Devel Ther. 2025. PMID: 39876987 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

