Drug-target identification in COVID-19 disease mechanisms using computational systems biology approaches
- PMID: 38414974
- PMCID: PMC10897000
- DOI: 10.3389/fimmu.2023.1282859
Drug-target identification in COVID-19 disease mechanisms using computational systems biology approaches
Abstract
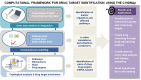
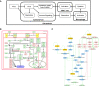
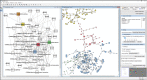
Introduction: The COVID-19 Disease Map project is a large-scale community effort uniting 277 scientists from 130 Institutions around the globe. We use high-quality, mechanistic content describing SARS-CoV-2-host interactions and develop interoperable bioinformatic pipelines for novel target identification and drug repurposing.
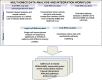
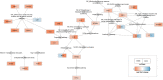
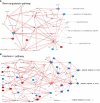
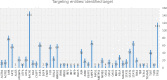
Methods: Extensive community work allowed an impressive step forward in building interfaces between Systems Biology tools and platforms. Our framework can link biomolecules from omics data analysis and computational modelling to dysregulated pathways in a cell-, tissue- or patient-specific manner. Drug repurposing using text mining and AI-assisted analysis identified potential drugs, chemicals and microRNAs that could target the identified key factors.
Results: Results revealed drugs already tested for anti-COVID-19 efficacy, providing a mechanistic context for their mode of action, and drugs already in clinical trials for treating other diseases, never tested against COVID-19.
Discussion: The key advance is that the proposed framework is versatile and expandable, offering a significant upgrade in the arsenal for virus-host interactions and other complex pathologies.
Keywords: SARS-CoV-2; disease maps; dynamic models; large-scale community effort; mechanistic models; systems biology; systems medicine.
Copyright © 2024 Niarakis, Ostaszewski, Mazein, Kuperstein, Kutmon, Gillespie, Funahashi, Acencio, Hemedan, Aichem, Klein, Czauderna, Burtscher, Yamada, Hiki, Hiroi, Hu, Pham, Ehrhart, Willighagen, Valdeolivas, Dugourd, Messina, Esteban-Medina, Peña-Chilet, Rian, Soliman, Aghamiri, Puniya, Naldi, Helikar, Singh, Fernández, Bermudez, Tsirvouli, Montagud, Noël, Ponce-de-Leon, Maier, Bauch, Gyori, Bachman, Luna, Piñero, Furlong, Balaur, Rougny, Jarosz, Overall, Phair, Perfetto, Matthews, Rex, Orlic-Milacic, Gomez, De Meulder, Ravel, Jassal, Satagopam, Wu, Golebiewski, Gawron, Calzone, Beckmann, Evelo, D’Eustachio, Schreiber, Saez-Rodriguez, Dopazo, Kuiper, Valencia, Wolkenhauer, Kitano, Barillot, Auffray, Balling, Schneider and the COVID-19 Disease Map Community.
Conflict of interest statement
AN collaborates with SANOFI-AVENTIS R&D via a public–private partnership grant CIFRE contract, n° 2020/0766. DM and AB are employed at Labvantage-Biomax GmbH and will be affected by any effect of this publication on the commercial version of the AILANI software. JB and BG received consulting fees from Two Six Labs, LLC. TH has served as a shareholder and has consulted for Discovery Collective, Inc. RB and RS are founders and shareholders of MEGENO SA and ITTM SA. JS-R reports funding from GSK, Pfizer and Sanofi and fees/honoraria from Travere Therapeutics, Stadapharm, Astex, Owkin, Pfizer and Grunenthal. JP and LF are employees and shareholders of MedBioinformatics Solutions SL. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

