Construction of a co-expression network affecting intramuscular fat content and meat color redness based on transcriptome analysis
- PMID: 38415055
- PMCID: PMC10897757
- DOI: 10.3389/fgene.2024.1351429
Construction of a co-expression network affecting intramuscular fat content and meat color redness based on transcriptome analysis
Abstract
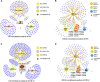
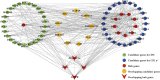
Introduction: Intramuscular fat content (IFC) and meat color are vital indicators of pork quality. Methods: A significant positive correlation between IFC and redness of meat color (CIE a* value) indicates that these two traits are likely to be regulated by shared molecular pathways.To identify candidate genes, hub genes, and signaling pathways that regulate these two traits, we measured the IFC and CIE a* value in 147 hybrid pigs, and selected individuls with extreme phenotypes for transcriptome analysis. Results: The results revealed 485 and 394 overlapping differentially expressed genes (DEGs), using the DESeq2, limma, and edgeR packages, affecting the IFC and CIE a* value, respectively. Weighted gene co-expression network analysis (WGCNA) identified four modules significantly correlated with the IFC and CIE a* value. Moreover, we integrated functional enrichment analysis results based on DEGs, GSEA, and WGCNA conditions to identify candidate genes, and identified 47 and 53 candidate genes affecting the IFC and CIE a* value, respectively. The protein protein interaction (PPI) network analysis of candidate genes showed that 5 and 13 hub genes affect the IFC and CIE a* value, respectively. These genes mainly participate in various pathways related to lipid metabolism and redox reactions. Notably, four crucial hub genes (MYC, SOX9, CEBPB, and PPAGRC1A) were shared for these two traits. Discussion and conclusion: After functional annotation of these four hub genes, we hypothesized that the SOX9/CEBPB/PPARGC1A axis could co-regulate lipid metabolism and the myoglobin redox response. Further research on these hub genes, especially the SOX9/CEBPB/PPARGC1A axis, will help to understand the molecular mechanism of the co-regulation of the IFC and CIE a* value, which will provide a theoretical basis for improving pork quality.
Keywords: RNA-seq; functional enrichment analysis; hub gene; intramuscular fat content; meat color redness.
Copyright © 2024 Wang, Hou, Yang, Men, Qi, Xu and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Muscle Transcriptome Analysis Reveals Potential Candidate Genes and Pathways Affecting Intramuscular Fat Content in Pigs.Front Genet. 2020 Aug 11;11:877. doi: 10.3389/fgene.2020.00877. eCollection 2020. Front Genet. 2020. PMID: 32849841 Free PMC article.
-
Prediction of hub genes associated with intramuscular fat content in Nelore cattle.BMC Genomics. 2019 Jun 25;20(1):520. doi: 10.1186/s12864-019-5904-x. BMC Genomics. 2019. PMID: 31238883 Free PMC article.
-
Identification of Key Modules and Hub Genes Involved in Regulating the Color of Chicken Breast Meat Using WGCNA.Animals (Basel). 2023 Jul 19;13(14):2356. doi: 10.3390/ani13142356. Animals (Basel). 2023. PMID: 37508133 Free PMC article.
-
Integration of transcriptome and machine learning to identify the potential key genes and regulatory networks affecting drip loss in pork.J Anim Sci. 2024 Jan 3;102:skae164. doi: 10.1093/jas/skae164. J Anim Sci. 2024. PMID: 38865489 Free PMC article.
-
Genome-wide association study reveals a quantitative trait locus and two candidate genes on Sus scrofa chromosome 5 affecting intramuscular fat content in Suhuai pigs.Animal. 2021 Sep;15(9):100341. doi: 10.1016/j.animal.2021.100341. Epub 2021 Aug 21. Animal. 2021. PMID: 34425484
References
LinkOut - more resources
Full Text Sources
Research Materials

