Exploring the secretome of Corynebacterium glutamicum ATCC 13032
- PMID: 38415189
- PMCID: PMC10896948
- DOI: 10.3389/fbioe.2024.1348184
Exploring the secretome of Corynebacterium glutamicum ATCC 13032
Abstract
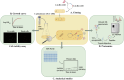
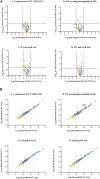
The demand for alternative sources of food proteins is increasing due to the limitations and challenges associated with conventional food production. Advances in biotechnology have enabled the production of proteins using microorganisms, thus prompting the exploration of attractive microbial hosts capable of producing functional proteins in high titers. Corynebacterium glutamicum is widely used in industry for the production of amino acids and has many advantages as a host organism for recombinant protein production. However, its performance in this area is limited by low yields of target proteins and high levels of native protein secretion. Despite representing a challenge for heterologous protein production, the C. glutamicum secretome has not been fully characterized. In this study, state-of-the-art mass spectrometry-based proteomics was used to identify and analyze the proteins secreted by C. glutamicum. Both the wild-type strain and a strain that produced and secreted a recombinant β-lactoglobulin protein were analyzed. A total of 427 proteins were identified in the culture supernatants, with 148 predicted to possess a secretion signal peptide. MS-based proteomics on the secretome enabled a comprehensive characterization and quantification (based on abundance) of the secreted proteins through label-free quantification (LFQ). The top 12 most abundant proteins accounted for almost 80% of the secretome. These are uncharacterized proteins of unknown function, resuscitation promoting factors, protein PS1, Porin B, ABC-type transporter protein and hypothetical membrane protein. The data can be leveraged for protein production by, e.g., utilizing the signal peptides of the most abundant proteins to improve secretion of heterologous proteins. In addition, secretory stress can potentially be alleviated by inactivating non-essential secreted proteins. Here we provide targets by identifying the most abundant, secreted proteins of which majority are of unknown function. The data from this study can thus provide valuable insight for researchers looking to improve protein secretion and optimize C. glutamicum as a host for secretory protein production.
Keywords: Corynebacterium glutamicum; recombinant protein production; secretome analysis; α-lactalbumin; β-lactoglobulin.
Copyright © 2024 Balasubramanian, Køhler, Jers, Jensen and Mijakovic.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Development of a new platform for secretory production of recombinant proteins in Corynebacterium glutamicum.Biotechnol Bioeng. 2016 Jan;113(1):163-72. doi: 10.1002/bit.25692. Epub 2015 Sep 8. Biotechnol Bioeng. 2016. PMID: 26134574
-
A secretion biosensor for monitoring Sec-dependent protein export in Corynebacterium glutamicum.Microb Cell Fact. 2020 Jan 21;19(1):11. doi: 10.1186/s12934-019-1273-z. Microb Cell Fact. 2020. PMID: 31964372 Free PMC article.
-
Protein secretion in Corynebacterium glutamicum.Crit Rev Biotechnol. 2017 Jun;37(4):541-551. doi: 10.1080/07388551.2016.1206059. Epub 2016 Oct 13. Crit Rev Biotechnol. 2017. PMID: 27737570 Review.
-
Development of a Novel Gene Expression System for Secretory Production of Heterologous Proteins via the General Secretory (Sec) Pathway in Corynebacterium Glutamicum.Iran J Biotechnol. 2018 Apr 18;16(1):e1746. doi: 10.21859/ijb.1746. eCollection 2018 Apr. Iran J Biotechnol. 2018. PMID: 30555839 Free PMC article.
-
Recombinant Protein Expression System in Corynebacterium glutamicum and Its Application.Front Microbiol. 2018 Oct 26;9:2523. doi: 10.3389/fmicb.2018.02523. eCollection 2018. Front Microbiol. 2018. PMID: 30416490 Free PMC article. Review.
References
-
- Balasubramanian S., Chen J., Wigneswaran V., Bang-Berthelsen C. H., Jensen P. R. (2021). Droplet-based microfluidic high throughput screening of Corynebacterium glutamicum for efficient heterologous protein production and secretion. Front. Bioeng. Biotechnol. 9, 668513. 10.3389/fbioe.2021.668513 - DOI - PMC - PubMed
-
- Baumgart M., Unthan S., Rückert C., Sivalingam J., Grünberger A., Kalinowski J., et al. (2013). Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl. Environ. Microbiol. 79, 6006–6015. 10.1128/AEM.01634-13 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

