Infection biology of Salmonella enterica
- PMID: 38415623
- PMCID: PMC11636313
- DOI: 10.1128/ecosalplus.esp-0001-2023
Infection biology of Salmonella enterica
Abstract
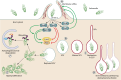
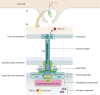
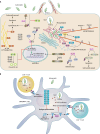
Salmonella enterica is the leading cause of bacterial foodborne illness in the USA, with an estimated 95% of salmonellosis cases due to the consumption of contaminated food products. Salmonella can cause several different disease syndromes, with the most common being gastroenteritis, followed by bacteremia and typhoid fever. Among the over 2,600 currently identified serotypes/serovars, some are mostly host-restricted and host-adapted, while the majority of serotypes can infect a broader range of host species and are associated with causing both livestock and human disease. Salmonella serotypes and strains within serovars can vary considerably in the severity of disease that may result from infection, with some serovars that are more highly associated with invasive disease in humans, while others predominantly cause mild gastroenteritis. These observed clinical differences may be caused by the genetic make-up and diversity of the serovars. Salmonella virulence systems are very complex containing several virulence-associated genes with different functions that contribute to its pathogenicity. The different clinical syndromes are associated with unique groups of virulence genes, and strains often differ in the array of virulence traits they display. On the chromosome, virulence genes are often clustered in regions known as Salmonella pathogenicity islands (SPIs), which are scattered throughout different Salmonella genomes and encode factors essential for adhesion, invasion, survival, and replication within the host. Plasmids can also carry various genes that contribute to Salmonella pathogenicity. For example, strains from several serovars associated with significant human disease, including Choleraesuis, Dublin, Enteritidis, Newport, and Typhimurium, can carry virulence plasmids with genes contributing to attachment, immune system evasion, and other roles. The goal of this comprehensive review is to provide key information on the Salmonella virulence, including the contributions of genes encoded in SPIs and plasmids during Salmonella pathogenesis.
Keywords: Salmonella; pathogenicity; virulence factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- CDC . 2019. Antibiotic resistance threats in the United States, 2019
-
- (IFSAC), T.I.F.S.A.C . 2019. Foodborne illness source attribution estimates for 2017 for Salmonella, Escherichia coli O157,Listeria monocytogenes, and Campylobacter using multi-year outbreak surveillance data,United States
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

