CSB and SMARCAL1 compete for RPA32 at stalled forks and differentially control the fate of stalled forks in BRCA2-deficient cells
- PMID: 38416570
- PMCID: PMC11109976
- DOI: 10.1093/nar/gkae154
CSB and SMARCAL1 compete for RPA32 at stalled forks and differentially control the fate of stalled forks in BRCA2-deficient cells
Abstract
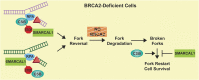
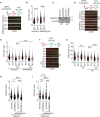
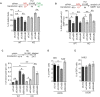
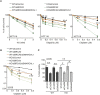
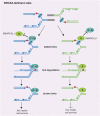
CSB (Cockayne syndrome group B) and SMARCAL1 (SWI/SNF-related, matrix-associated, actin-dependent, regulator of chromatin, subfamily A-like 1) are DNA translocases that belong to the SNF2 helicase family. They both are enriched at stalled replication forks. While SMARCAL1 is recruited by RPA32 to stalled forks, little is known about whether RPA32 also regulates CSB's association with stalled forks. Here, we report that CSB directly interacts with RPA, at least in part via a RPA32C-interacting motif within the N-terminal region of CSB. Modeling of the CSB-RPA32C interaction suggests that CSB binds the RPA32C surface previously shown to be important for binding of UNG2 and SMARCAL1. We show that this interaction is necessary for promoting fork slowing and fork degradation in BRCA2-deficient cells but dispensable for mediating restart of stalled forks. CSB competes with SMARCAL1 for RPA32 at stalled forks and acts non-redundantly with SMARCAL1 to restrain fork progression in response to mild replication stress. In contrast to CSB stimulated restart of stalled forks, SMARCAL1 inhibits restart of stalled forks in BRCA2-deficient cells, likely by suppressing BIR-mediated repair of collapsed forks. Loss of CSB leads to re-sensitization of SMARCAL1-depleted BRCA2-deficient cells to chemodrugs, underscoring a role of CSB in targeted cancer therapy.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Cockayne syndrome group B protein regulates fork restart, fork progression and MRE11-dependent fork degradation in BRCA1/2-deficient cells.Nucleic Acids Res. 2021 Dec 16;49(22):12836-12854. doi: 10.1093/nar/gkab1173. Nucleic Acids Res. 2021. PMID: 34871413 Free PMC article.
-
Cockayne syndrome group B protein uses its DNA translocase activity to promote mitotic DNA synthesis.DNA Repair (Amst). 2022 Aug;116:103354. doi: 10.1016/j.dnarep.2022.103354. Epub 2022 Jun 17. DNA Repair (Amst). 2022. PMID: 35738143
-
Identification and characterization of SMARCAL1 protein complexes.PLoS One. 2013 May 9;8(5):e63149. doi: 10.1371/journal.pone.0063149. Print 2013. PLoS One. 2013. PMID: 23671665 Free PMC article.
-
The role of SMARCAL1 in replication fork stability and telomere maintenance.DNA Repair (Amst). 2017 Aug;56:129-134. doi: 10.1016/j.dnarep.2017.06.015. Epub 2017 Jun 10. DNA Repair (Amst). 2017. PMID: 28623093 Review.
-
Role of Cockayne Syndrome Group B Protein in Replication Stress: Implications for Cancer Therapy.Int J Mol Sci. 2022 Sep 6;23(18):10212. doi: 10.3390/ijms231810212. Int J Mol Sci. 2022. PMID: 36142121 Free PMC article. Review.
Cited by
-
The Ski2 helicase ASCC3 unwinds DNA upon fork stalling to control replication stress responses.bioRxiv [Preprint]. 2025 Jul 26:2025.07.24.666583. doi: 10.1101/2025.07.24.666583. bioRxiv. 2025. PMID: 40777259 Free PMC article. Preprint.
-
p53-dependent crosstalk between DNA replication integrity and redox metabolism mediated through a NRF2-PARP1 axis.Nucleic Acids Res. 2024 Nov 11;52(20):12351-12377. doi: 10.1093/nar/gkae811. Nucleic Acids Res. 2024. PMID: 39315696 Free PMC article.
References
-
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. - PubMed
-
- Halazonetis T.D., Gorgoulis V.G., Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008; 319:1352–1355. - PubMed
-
- Macheret M., Halazonetis T. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015; 10:425–448. - PubMed
-
- Gaillard H., García-Muse T., Aguilera A. Replication stress and cancer. Nat. Rev. Cancer. 2015; 15:276–289. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

