Respiratory disease complex due to mixed viral infections in chicken in Jordan
- PMID: 38417340
- PMCID: PMC10907842
- DOI: 10.1016/j.psj.2024.103565
Respiratory disease complex due to mixed viral infections in chicken in Jordan
Abstract
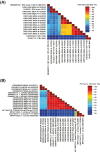
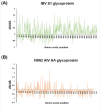
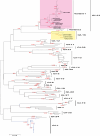
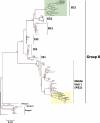
The global distribution of avian respiratory viruses highlights the need for effective surveillance programs and international collaboration to monitor viral circulation and implement timely control measures. In the current study, we aim to provide a comprehensive overview of avian respiratory viral infections in the poultry flocks in Jordan, focusing on the major viruses involved, their epidemiology, clinical manifestations, and evolution based on viroinformatics that will be helpful to improve the diagnostic methods, and control strategies including vaccines in the region. In this research, various poultry broiler groups in Jordan experiencing respiratory symptoms were tested for respiratory viral pathogens from January 2021 to February 2022. The mortality rates observed in the examined groups varied between 6% and 40%. The identified strains were authenticated using the RT-qPCR assay. Furthermore, they underwent in-depth characterisation through the sequencing of the complete spike (S1) gene for infectious bronchitis virus (IBV) and the haemagglutinin (HA) gene for avian influenza virus (AIV) subtype H9N2. Co-infection of IBV and AIV H9N2 viruses was detected through molecular analysis. The IBV strains showed affiliation with the variant groups GI-16 (3 strains) and GI-23 (9 strains) and exhibited numerous mutations. Meanwhile, H9N2 avian influenza viruses displayed various changes in amino acids within the HA gene, suggesting the influence of antibody-driven selection pressure. The phylogenetic analysis revealed that the H9N2 viruses identified in this investigation shared close genetic ties with EG3 (3 strains) and the Middle East group (ME1; 8 strains). These strains have been recently found in Jordan and nearby countries in the Middle East. Moreover, their HA genes exhibited similarities to viruses belonging to the G1-like lineage. In conclusion, avian respiratory viral infections remain a significant concern for the poultry industry, requiring constant vigilance and proactive measures to minimise their impact. Continued surveillance, robust diagnostic methods, effective vaccines, and international cooperation are essential components of a comprehensive approach to combat avian respiratory viral infections (AI, IBV, ND and ILT 'viruses) and safeguard avian health and global poultry production.
Keywords: Jordan; avian influenza viruses; infectious bronchitis virus; poultry health; respiratory infection.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Prevalence of avian respiratory viruses in broiler flocks in Egypt.Poult Sci. 2016 Jun 1;95(6):1271-80. doi: 10.3382/ps/pew068. Epub 2016 Mar 14. Poult Sci. 2016. PMID: 26976895 Free PMC article.
-
Surveillance and Coinfection Dynamics of Infectious Bronchitis Virus and Avian Influenza H9N2 in Moroccan Broiler Farms (2021-2023): Phylogenetic Insights and Impact on Poultry Health.Viruses. 2025 May 30;17(6):786. doi: 10.3390/v17060786. Viruses. 2025. PMID: 40573377 Free PMC article.
-
Predominance of low pathogenic avian influenza virus H9N2 in the respiratory co-infections in broilers in Tunisia: a longitudinal field study, 2018-2020.Vet Res. 2023 Oct 3;54(1):88. doi: 10.1186/s13567-023-01204-7. Vet Res. 2023. PMID: 37789451 Free PMC article.
-
Review analysis and impact of co-circulating H5N1 and H9N2 avian influenza viruses in Bangladesh.Epidemiol Infect. 2018 Jul;146(10):1259-1266. doi: 10.1017/S0950268818001292. Epub 2018 May 21. Epidemiol Infect. 2018. PMID: 29781424 Free PMC article.
-
A brief summary of the epidemiology and genetic relatedness of avian influenza H9N2 virus in birds and mammals in the Middle East and North Africa.Epidemiol Infect. 2017 Dec;145(16):3320-3333. doi: 10.1017/S0950268817002576. Epub 2017 Nov 23. Epidemiol Infect. 2017. PMID: 29168447 Free PMC article. Review.
References
-
- Al-Garib S., Agha A., Al-Mesilaty L. Low pathogenic avian influenza H9N2: world-wide distribution. J. World's Poult. Sci. 2016;72:125–136.
-
- Al-Natour M.Q., Abo-Shehada M.N. Sero-prevalence of avian influenza among broiler-breeder flocks in Jordan. Prev. Vet. Med. 2005;70:45–50. - PubMed
-
- Bashashati M., Vasfi Marandi M., Sabouri F. Genetic diversity of early (1998) and recent 2010. Avian influenza H9N2 virus strains isolated from poultry in Iran. Arch Virol. 2013;158:2089–2100. - PubMed
-
- Cavanagh D., Davis P., Cook J. Infectious bronchitis virus: evidence for recombination within the Massachusetts serotype. Avian Pathol. 1992;21:401–408. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

