Functional analysis and clinical classification of 462 germline BRCA2 missense variants affecting the DNA binding domain
- PMID: 38417439
- PMCID: PMC10940015
- DOI: 10.1016/j.ajhg.2024.02.002
Functional analysis and clinical classification of 462 germline BRCA2 missense variants affecting the DNA binding domain
Abstract
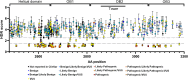
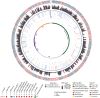
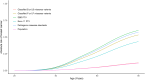
Variants of uncertain significance (VUSs) in BRCA2 are a common result of hereditary cancer genetic testing. While more than 4,000 unique VUSs, comprised of missense or intronic variants, have been identified in BRCA2, the few missense variants now classified clinically as pathogenic or likely pathogenic are predominantly located in the region encoding the C-terminal DNA binding domain (DBD). We report on functional evaluation of the influence of 462 BRCA2 missense variants affecting the DBD on DNA repair activity of BRCA2 using a homology-directed DNA double-strand break repair assay. Of these, 137 were functionally abnormal, 313 were functionally normal, and 12 demonstrated intermediate function. Comparisons with other functional studies of BRCA2 missense variants yielded strong correlations. Sequence-based in silico prediction models had high sensitivity, but limited specificity, relative to the homology-directed repair assay. Combining the functional results with clinical and genetic data in an American College of Medical Genetics (ACMG)/Association for Molecular Pathology (AMP)-like variant classification framework from a clinical testing laboratory, after excluding known splicing variants and functionally intermediate variants, classified 431 of 442 (97.5%) missense variants (129 as pathogenic/likely pathogenic and 302 as benign/likely benign). Functionally abnormal variants classified as pathogenic by ACMG/AMP rules were associated with a slightly lower risk of breast cancer (odds ratio [OR] 5.15, 95% confidence interval [CI] 3.43-7.83) than BRCA2 DBD protein truncating variants (OR 8.56, 95% CI 6.03-12.36). Overall, functional studies of BRCA2 variants using validated assays substantially improved the variant classification yield from ACMG/AMP models and are expected to improve clinical management of many individuals found to harbor germline BRCA2 missense VUS.
Keywords: BRCA2; HDR; VUS; cancer; predisposition genes; variant classification.
Copyright © 2024 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.K., T.P., J.D.W., C.M.C., C.A.S., C.C.Y., K.F., Z.H., and M.E.R. are employees of Ambry Genetics Inc. All other authors declare no competing interests.
Figures


References
-
- Goldgar D.E., Easton D.F., Deffenbaugh A.M., Monteiro A.N.A., Tavtigian S.V., Couch F.J., Breast Cancer Information Core BIC Steering Committee Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am. J. Hum. Genet. 2004;75:535–544. - PMC - PubMed
-
- Parsons M.T., Tudini E., Li H., Hahnen E., Wappenschmidt B., Feliubadaló L., Aalfs C.M., Agata S., Aittomäki K., Alducci E., et al. Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum. Mutat. 2019;40:1557–1578. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

