Flowering time: From physiology, through genetics to mechanism
- PMID: 38417841
- PMCID: PMC11060688
- DOI: 10.1093/plphys/kiae109
Flowering time: From physiology, through genetics to mechanism
Abstract
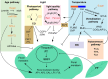
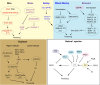
Plant species have evolved different requirements for environmental/endogenous cues to induce flowering. Originally, these varying requirements were thought to reflect the action of different molecular mechanisms. Thinking changed when genetic and molecular analysis in Arabidopsis thaliana revealed that a network of environmental and endogenous signaling input pathways converge to regulate a common set of "floral pathway integrators." Variation in the predominance of the different input pathways within a network can generate the diversity of requirements observed in different species. Many genes identified by flowering time mutants were found to encode general developmental and gene regulators, with their targets having a specific flowering function. Studies of natural variation in flowering were more successful at identifying genes acting as nodes in the network central to adaptation and domestication. Attention has now turned to mechanistic dissection of flowering time gene function and how that has changed during adaptation. This will inform breeding strategies for climate-proof crops and help define which genes act as critical flowering nodes in many other species.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

