Paired single-B-cell transcriptomics and receptor sequencing reveal activation states and clonal signatures that characterize B cells in acute myeloid leukemia
- PMID: 38418394
- PMCID: PMC10910691
- DOI: 10.1136/jitc-2023-008318
Paired single-B-cell transcriptomics and receptor sequencing reveal activation states and clonal signatures that characterize B cells in acute myeloid leukemia
Abstract
Background: Acute myeloid leukemia (AML) is associated with a dismal prognosis. Immune checkpoint blockade (ICB) to induce antitumor activity in AML patients has yielded mixed results. Despite the pivotal role of B cells in antitumor immunity, a comprehensive assessment of B lymphocytes within AML's immunological microenvironment along with their interaction with ICB remains rather constrained.
Methods: We performed an extensive analysis that involved paired single-cell RNA and B-cell receptor (BCR) sequencing on 52 bone marrow aspirate samples. These samples included 6 from healthy bone marrow donors (normal), 24 from newly diagnosed AML patients (NewlyDx), and 22 from 8 relapsed or refractory AML patients (RelRef), who underwent assessment both before and after azacitidine/nivolumab treatment.
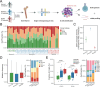
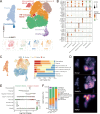
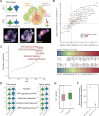
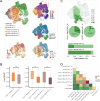
Results: We delineated nine distinct subtypes of B cell lineage in the bone marrow. AML patients exhibited reduced nascent B cell subgroups but increased differentiated B cells compared with healthy controls. The limited diversity of BCR profiles and extensive somatic hypermutation indicated antigen-driven affinity maturation within the tumor microenvironment of RelRef patients. We established a strong connection between the activation or stress status of naïve and memory B cells, as indicated by AP-1 activity, and their differentiation state. Remarkably, atypical memory B cells functioned as specialized antigen-presenting cells closely interacting with AML malignant cells, correlating with AML stemness and worse clinical outcomes. In the AML microenvironment, plasma cells demonstrated advanced differentiation and heightened activity. Notably, the clinical response to ICB was associated with B cell clonal expansion and plasma cell function.
Conclusions: Our findings establish a comprehensive framework for profiling the phenotypic diversity of the B cell lineage in AML patients, while also assessing the implications of immunotherapy. This will serve as a valuable guide for future inquiries into AML treatment strategies.
Keywords: Computational Biology; Gene Expression Profiling; Immune Checkpoint Inhibitors; Tumor Microenvironment.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Figures


References
-
- Kolb HJ, Schattenberg A, Goldman JM, et al. . Graft-versus-leukemia effect of donor lymphocyte transfusions in marrow grafted patients. Blood 1995;86:2041–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
