Molecular and clinical analyses of PHF6 mutant myeloid neoplasia provide their pathogenesis and therapeutic targeting
- PMID: 38418452
- PMCID: PMC10901781
- DOI: 10.1038/s41467-024-46134-w
Molecular and clinical analyses of PHF6 mutant myeloid neoplasia provide their pathogenesis and therapeutic targeting
Abstract
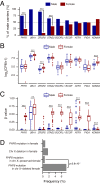
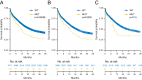
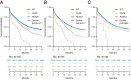
PHF6 mutations (PHF6MT) are identified in various myeloid neoplasms (MN). However, little is known about the precise function and consequences of PHF6 in MN. Here we show three main findings in our comprehensive genomic and proteomic study. Firstly, we show a different pattern of genes correlating with PHF6MT in male and female cases. When analyzing male and female cases separately, in only male cases, RUNX1 and U2AF1 are co-mutated with PHF6. In contrast, female cases reveal co-occurrence of ASXL1 mutations and X-chromosome deletions with PHF6MT. Next, proteomics analysis reveals a direct interaction between PHF6 and RUNX1. Both proteins co-localize in active enhancer regions that define the context of lineage differentiation. Finally, we demonstrate a negative prognostic role of PHF6MT, especially in association with RUNX1. The negative effects on survival are additive as PHF6MT cases with RUNX1 mutations have worse outcomes when compared to cases carrying single mutation or wild-type.
© 2024. The Author(s).
Conflict of interest statement
This work was supported by an R35HL135795 (to J.P.M.), The Leukemia & Lymphoma Society TRP Award 6645-22 (to J.P.M.), and by a grant from the Edward P. Evans Foundation (to C.G.). The remaining authors declare no competing interests.
Figures
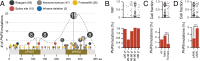


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

