Cryo-EM structure of cell-free synthesized human histamine 2 receptor/Gs complex in nanodisc environment
- PMID: 38418462
- PMCID: PMC10901899
- DOI: 10.1038/s41467-024-46096-z
Cryo-EM structure of cell-free synthesized human histamine 2 receptor/Gs complex in nanodisc environment
Abstract
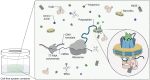
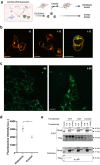
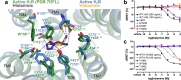
Here we describe the cryo-electron microscopy structure of the human histamine 2 receptor (H2R) in an active conformation with bound histamine and in complex with Gs heterotrimeric protein at an overall resolution of 3.4 Å. The complex was generated by cotranslational insertion of the receptor into preformed nanodisc membranes using cell-free synthesis in E. coli lysates. Structural comparison with the inactive conformation of H2R and the inactive and Gq-coupled active state of H1R together with structure-guided functional experiments reveal molecular insights into the specificity of ligand binding and G protein coupling for this receptor family. We demonstrate lipid-modulated folding of cell-free synthesized H2R, its agonist-dependent internalization and its interaction with endogenously synthesized H1R and H2R in HEK293 cells by applying a recently developed nanotransfer technique.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




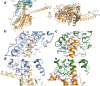
Similar articles
-
Specific Engineered G Protein Coupling to Histamine Receptors Revealed from Cellular Assay Experiments and Accelerated Molecular Dynamics Simulations.Int J Mol Sci. 2021 Sep 17;22(18):10047. doi: 10.3390/ijms221810047. Int J Mol Sci. 2021. PMID: 34576210 Free PMC article.
-
Cross-desensitization and cointernalization of H1 and H2 histamine receptors reveal new insights into histamine signal integration.Mol Pharmacol. 2013 May;83(5):1087-98. doi: 10.1124/mol.112.083394. Epub 2013 Mar 5. Mol Pharmacol. 2013. PMID: 23462507 Free PMC article.
-
The human histamine H2-receptor couples more efficiently to Sf9 insect cell Gs-proteins than to insect cell Gq-proteins: limitations of Sf9 cells for the analysis of receptor/Gq-protein coupling.J Neurochem. 2002 Feb;80(4):678-96. doi: 10.1046/j.0022-3042.2001.00746.x. J Neurochem. 2002. PMID: 11841575
-
H2 antihistamines: May be useful for combination therapies in cancer?Biochem Pharmacol. 2024 May;223:116164. doi: 10.1016/j.bcp.2024.116164. Epub 2024 Mar 24. Biochem Pharmacol. 2024. PMID: 38531422 Review.
-
Histamine H2 receptor radioligands: triumphs and challenges.Future Med Chem. 2021 Jun;13(12):1073-1081. doi: 10.4155/fmc-2021-0058. Epub 2021 Apr 28. Future Med Chem. 2021. PMID: 33906421 Review.
Cited by
-
Decoding ligand recognition and constitutive activation of histamine H3 and H4 receptors.Acta Pharmacol Sin. 2025 Aug 28. doi: 10.1038/s41401-025-01633-4. Online ahead of print. Acta Pharmacol Sin. 2025. PMID: 40877594
-
Cryo-EM Structures and AlphaFold3 Models of Histamine Receptors Reveal Diverse Ligand Binding and G Protein Bias.Pharmaceuticals (Basel). 2025 Feb 21;18(3):292. doi: 10.3390/ph18030292. Pharmaceuticals (Basel). 2025. PMID: 40143071 Free PMC article.
-
In-Cell DEER Spectroscopy of Nanodisc-Delivered Membrane Proteins in Living Cell Membranes.JACS Au. 2024 Sep 24;4(10):3766-3770. doi: 10.1021/jacsau.4c00702. eCollection 2024 Oct 28. JACS Au. 2024. PMID: 39483229 Free PMC article.
-
Structural insights into ligand recognition and G protein preferences across histamine receptors.Commun Biol. 2025 Jun 27;8(1):957. doi: 10.1038/s42003-025-08363-7. Commun Biol. 2025. PMID: 40579541 Free PMC article.
-
Cell-free protein synthesis platforms for accelerating drug discovery.Biotechnol Notes. 2025 Feb 19;6:126-132. doi: 10.1016/j.biotno.2025.02.001. eCollection 2025. Biotechnol Notes. 2025. PMID: 40123759 Free PMC article. Review.
References
-
- Delvalle J, Wang L, Gantz I, Yamada T. Characterization of H2 histamine receptor: linkage to both adenylate cyclase and [Ca2+]i signaling systems. Am. J. Physiol. -Gastr. L. 1992;263:G967–G972. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

