Proteomics-based vaccine targets annotation and design of multi-epitope vaccine against antibiotic-resistant Streptococcus gallolyticus
- PMID: 38418560
- PMCID: PMC10901886
- DOI: 10.1038/s41598-024-55372-3
Proteomics-based vaccine targets annotation and design of multi-epitope vaccine against antibiotic-resistant Streptococcus gallolyticus
Abstract
Streptococcus gallolyticus is a non-motile, gram-positive bacterium that causes infective endocarditis. S. gallolyticus has developed resistance to existing antibiotics, and no vaccine is currently available. Therefore, it is essential to develop an effective S. gallolyticus vaccine. Core proteomics was used in this study together with subtractive proteomics and reverse vaccinology approach to find antigenic proteins that could be utilized for the design of the S. gallolyticus multi-epitope vaccine. The pipeline identified two antigenic proteins as potential vaccine targets: penicillin-binding protein and the ATP synthase subunit. T and B cell epitopes from the specific proteins were forecasted employing several immunoinformatics and bioinformatics resources. A vaccine (360 amino acids) was created using a combination of seven cytotoxic T cell lymphocyte (CTL), three helper T cell lymphocyte (HTL), and five linear B cell lymphocyte (LBL) epitopes. To increase immune responses, the vaccine was paired with a cholera enterotoxin subunit B (CTB) adjuvant. The developed vaccine was highly antigenic, non-allergenic, and stable for human use. The vaccine's binding affinity and molecular interactions with the human immunological receptor TLR4 were studied using molecular mechanics/generalized Born surface area (MMGBSA), molecular docking, and molecular dynamic (MD) simulation analyses. Escherichia coli (strain K12) plasmid vector pET-28a ( +) was used to examine the ability of the vaccine to be expressed. According to the outcomes of these computer experiments, the vaccine is quite promising in terms of developing a protective immunity against diseases. However, in vitro and animal research are required to validate our findings.
Keywords: Immunoinformatics; Multi-epitope vaccine; Pan-genome; Reverse vaccinology.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

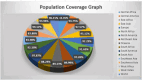






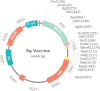
Similar articles
-
Prioritization of potential vaccine candidates and designing a multiepitope-based subunit vaccine against multidrug-resistant Salmonella Typhi str. CT18: A subtractive proteomics and immunoinformatics approach.Microb Pathog. 2021 Oct;159:105150. doi: 10.1016/j.micpath.2021.105150. Epub 2021 Aug 20. Microb Pathog. 2021. PMID: 34425197
-
Pan-vaccinomics strategy for developing a universal multi-epitope vaccine against endocarditis-related pathogens.Front Immunol. 2025 Apr 11;16:1524128. doi: 10.3389/fimmu.2025.1524128. eCollection 2025. Front Immunol. 2025. PMID: 40292293 Free PMC article.
-
Revolutionizing and identifying novel drug targets in Citrobacter koseri via subtractive proteomics and development of a multi-epitope vaccine using reverse vaccinology and immuno-informatics.J Biomol Struct Dyn. 2024 Feb 26:1-14. doi: 10.1080/07391102.2024.2316762. Online ahead of print. J Biomol Struct Dyn. 2024. PMID: 38407210
-
Immunogenic multi-epitope-based vaccine development to combat cyclosporiasis of immunocompromised patients applying computational biology method.Exp Parasitol. 2023 May;248:108497. doi: 10.1016/j.exppara.2023.108497. Epub 2023 Mar 10. Exp Parasitol. 2023. PMID: 36906252 Review.
-
The Promising Potential of Reverse Vaccinology-Based Next-Generation Vaccine Development over Conventional Vaccines against Antibiotic-Resistant Bacteria.Vaccines (Basel). 2023 Jul 20;11(7):1264. doi: 10.3390/vaccines11071264. Vaccines (Basel). 2023. PMID: 37515079 Free PMC article. Review.
Cited by
-
Reverse vaccinology and immunoinformatics approaches for multi-epitope vaccine design against Klebsiella pneumoniae reveal a novel vaccine target protein.J Genet Eng Biotechnol. 2025 Sep;23(3):100510. doi: 10.1016/j.jgeb.2025.100510. Epub 2025 May 23. J Genet Eng Biotechnol. 2025. PMID: 40854630 Free PMC article.
-
In Silico Subtractive Proteome Analysis to Design Multi-Epitope-Based Subunit Vaccine against Eikenella corrodens.J Microbiol Biotechnol. 2024 Nov 25;35:e2410015. doi: 10.4014/jmb.2410.10015. J Microbiol Biotechnol. 2024. PMID: 39809513 Free PMC article.
-
In silico design of peptide inhibitors for Dengue virus to treat Dengue virus-associated infections.Sci Rep. 2024 Jun 7;14(1):13130. doi: 10.1038/s41598-024-63064-1. Sci Rep. 2024. PMID: 38849372 Free PMC article.
-
Multi-epitope vaccines: a promising strategy against viral diseases in swine.Front Cell Infect Microbiol. 2024 Dec 20;14:1497580. doi: 10.3389/fcimb.2024.1497580. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39760092 Free PMC article. Review.
-
In Silico Design, Optimization, and Evaluation of a Multi-Epitope Vaccine Targeting the Clostridium perfringens Collagen Adhesin Protein.Microorganisms. 2025 May 16;13(5):1147. doi: 10.3390/microorganisms13051147. Microorganisms. 2025. PMID: 40431317 Free PMC article.
References
-
- Baddour LM, et al. Infective endocarditis: diagnosis, antimicrobial therapy, and management of complications: A statement for healthcare professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, and the Councils on Clinical Cardiology, Stroke, and Cardiovascular Surgery and Anesthesia, American Heart Association: endorsed by the Infectious Diseases Society of America. Circulation. 2005;111(23):e394–e434. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

