Untargeted metabolomic, and proteomic analysis identifies metabolic biomarkers and pathway alterations in individuals with 22q11.2 deletion syndrome
- PMID: 38418685
- PMCID: PMC10901937
- DOI: 10.1007/s11306-024-02088-0
Untargeted metabolomic, and proteomic analysis identifies metabolic biomarkers and pathway alterations in individuals with 22q11.2 deletion syndrome
Abstract
Introduction: The chromosome 22q11.2 deletion syndrome (22q11.2DS) is characterized by a well-defined microdeletion and is associated with a wide range of brain-related phenotypes including schizophrenia spectrum disorders (SCZ), autism spectrum disorders (ASD), anxiety disorders and attention deficit disorders (ADHD). The typically deleted region in 22q11.2DS contains multiple genes which haploinsufficiency has the potential of altering the protein and the metabolic profiles.
Objectives: Alteration in metabolic processes and downstream protein pathways during the early brain development may help to explain the increased prevalence of the observed neurodevelopmental phenotypes in 22q11.2DS. However, relatively little is known about the correlation of dysregulated protein/metabolite expression and neurobehavioral impairments in individuals who developed them over time.
Methods: In this study, we performed untargeted metabolic and proteomic analysis in plasma samples derived from 30 subjects including 16 participants with 22q11.2DS and 14 healthy controls (TD) enrolled in a longitudinal study, aiming to identify a metabolic and protein signature informing about the underlying mechanisms involved in disease development and progression. The metabolic and proteomic profiles were also compared between the participants with 22q11.2DS with and without various comorbidities, such as medical involvement, psychiatric conditions, and autism spectrum disorder (ASD) to detect potential changes among multiple specimens, collected overtime, with the aim to understand the basic underlying mechanisms involved in disease development and progression.
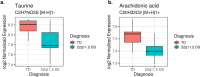
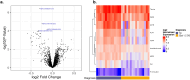
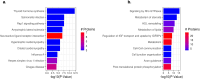
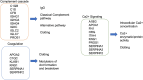
Results: We observed a large number of statistically significant differences in metabolites between the two groups. Among them, the levels of taurine and arachidonic acid were significantly lower in 22q11.2DS compared to the TD group. In addition, we identified 16 proteins that showed significant changes in expression levels (adjusted P < 0.05) in 22q11.2DS as compared to TD, including those involved in 70 pathways such as gene expression, the PI3K-Akt signaling pathway and the complement system. Within participants with 22q11.2DS, no significant changes in those with and without medical or psychiatric conditions were observed.
Conclusion: To our knowledge, this is the first report on plasma metabolic and proteomic profiling and on the identification of unique biomarkers in 22q11.2DS. These findings may suggest the potential role of the identified metabolites and proteins as biomarkers for the onset of comorbid conditions in 22q11.2DS. Ultimately, the altered protein pathways in 22q11.2DS may provide insights of the biological mechanisms underlying the neurodevelopmental phenotype and may provide missing molecular outcome measures in future clinical trials to assess early-diagnosis treatment and the efficacy of response to targeted treatment.
Keywords: 22q11.2 deletion syndrome; APS; AS; Biomarker; Metabolomics; Pathways; Proteomics.
© 2024. The Author(s).
Conflict of interest statement
M.Z.: No disclosures to report; K. A: No disclosures to report; A.Q. & S.Y.: AQ and SY are co-founders of, are employed by, and own stock in Dalton Bioanalytics Inc.; B.J.: No disclosures to report; H.B.: H. B. is an employee of Epistemic AI; F.T.: No disclosures to report.
Figures


Similar articles
-
Untargeted metabolic analysis in dried blood spots reveals metabolic signature in 22q11.2 deletion syndrome.Transl Psychiatry. 2022 Mar 9;12(1):97. doi: 10.1038/s41398-022-01859-4. Transl Psychiatry. 2022. PMID: 35264571 Free PMC article.
-
Deep psychophysiological phenotyping of adolescents and adults with 22q11.2 deletion syndrome: a multilevel approach to defining core disease processes.BMC Psychiatry. 2023 Jun 13;23(1):425. doi: 10.1186/s12888-023-04888-5. BMC Psychiatry. 2023. PMID: 37312091 Free PMC article.
-
22q11.2 duplication syndrome: elevated rate of autism spectrum disorder and need for medical screening.Mol Autism. 2016 May 6;7:27. doi: 10.1186/s13229-016-0090-z. eCollection 2016. Mol Autism. 2016. PMID: 27158440 Free PMC article.
-
Developmental trajectories in 22q11.2 deletion.Am J Med Genet C Semin Med Genet. 2015 Jun;169(2):172-81. doi: 10.1002/ajmg.c.31435. Epub 2015 May 18. Am J Med Genet C Semin Med Genet. 2015. PMID: 25989227 Free PMC article. Review.
-
[Neurocognitive and psychiatric management of the 22q11.2 deletion syndrome].Encephale. 2015 Jun;41(3):266-73. doi: 10.1016/j.encep.2014.10.005. Epub 2014 Dec 16. Encephale. 2015. PMID: 25523123 Review. French.
Cited by
-
Exploring the Association Between Human Blood Metabolites and Autism Spectrum Disorder Risk: A Bidirectional Mendelian Randomization Study.Health Sci Rep. 2025 Mar 3;8(3):e70528. doi: 10.1002/hsr2.70528. eCollection 2025 Mar. Health Sci Rep. 2025. PMID: 40041792 Free PMC article.
-
Brain and behavioural anomalies caused by Tbx1 haploinsufficiency are corrected by vitamin B12.Life Sci Alliance. 2024 Nov 20;8(2):e202403075. doi: 10.26508/lsa.202403075. Print 2025 Feb. Life Sci Alliance. 2024. PMID: 39567195 Free PMC article.
References
-
- Almeida N, Rodriguez J, Parada IP, Perez-Riverol Y, Woldmar N, Kim Y, Oskolas H, Betancourt L, Valdés JG, Barbara Sahlin K, Luciana Pizzatti A, Szasz M, Kárpáti S, Appelqvist R, Malm J, Domont GB, Nogueira FCS, Marko-Varga G, Sanchez A. Mapping the melanoma plasma proteome (MPP) using single-shot proteomics interfaced with the WiMT database. Cancers. 2021 doi: 10.3390/cancers13246224. - DOI - PMC - PubMed
-
- Alshweki A, Muñuzuri AP, Baña AM, José de Castro M, Andrade F, Aldamiz-Echevarría L, Sáenz M, de Pipaón JM, Fraga ML, Couce, Effects of different arachidonic acid supplementation on psychomotor development in very preterm infants; a randomized controlled trial. Nutrition Journal. 2015;14:101. doi: 10.1186/s12937-015-0091-3. - DOI - PMC - PubMed
-
- Antshel KM, Kates WR, Roizen N, Fremont W, Shprintzen RJ. 22q11.2 deletion syndrome: genetics, neuroanatomy and cognitive/behavioral features keywords. Child Neuropsychology: A Journal on Normal and Abnormal Development in Childhood and Adolescence. 2005;11(1):5–19. doi: 10.1080/09297040590911185. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
