SOX17 enables immune evasion of early colorectal adenomas and cancers
- PMID: 38418875
- PMCID: PMC11969226
- DOI: 10.1038/s41586-024-07135-3
SOX17 enables immune evasion of early colorectal adenomas and cancers
Abstract
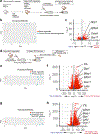
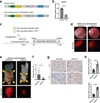
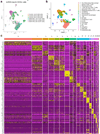
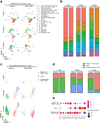
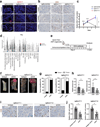
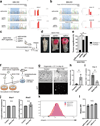
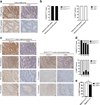
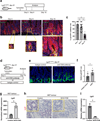
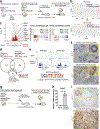
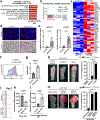
A hallmark of cancer is the avoidance of immune destruction. This process has been primarily investigated in locally advanced or metastatic cancer1-3; however, much less is known about how pre-malignant or early invasive tumours evade immune detection. Here, to understand this process in early colorectal cancers (CRCs), we investigated how naive colon cancer organoids that were engineered in vitro to harbour Apc-null, KrasG12D and Trp53-null (AKP) mutations adapted to the in vivo native colonic environment. Comprehensive transcriptomic and chromatin analyses revealed that the endoderm-specifying transcription factor SOX17 became strongly upregulated in vivo. Notably, whereas SOX17 loss did not affect AKP organoid propagation in vitro, its loss markedly reduced the ability of AKP tumours to persist in vivo. The small fraction of SOX17-null tumours that grew displayed notable interferon-γ (IFNγ)-producing effector-like CD8+ T cell infiltrates in contrast to the immune-suppressive microenvironment in wild-type counterparts. Mechanistically, in both endogenous Apc-null pre-malignant adenomas and transplanted organoid-derived AKP CRCs, SOX17 suppresses the ability of tumour cells to sense and respond to IFNγ, preventing anti-tumour T cell responses. Finally, SOX17 engages a fetal intestinal programme that drives differentiation away from LGR5+ tumour cells to produce immune-evasive LGR5- tumour cells with lower expression of major histocompatibility complex class I (MHC-I). We propose that SOX17 is a transcription factor that is engaged during the early steps of colon cancer to orchestrate an immune-evasive programme that permits CRC initiation and progression.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
Ö.H.Y. holds equity and is a SAB member in Ava Lifesciences and AI Proteins. Ö.H.Y. receives research support from Microbial Machines. Ö.H.Y. is a consultant for Nestle. T.J. is a member of the Board of Directors of Amgen and Thermo Fisher Scientific and a co-founder of Dragonfly Therapeutics and T2 Biosystems; serves on the Scientific Advisory Board of Dragonfly Therapeutics, SQZ Biotech, and Skyhawk Therapeutics; and is President of Break Through Cancer. T.J.’s laboratory currently receives funding from Johnson & Johnson and The Lustgarten Foundation, but these funds did not support the research described in this manuscript. None of the other authors have any conflicts to declare.
Figures









Comment in
-
SOX17 orchestrates immune evasion in early colorectal adenomas and cancers.Immunology. 2024 Oct;173(2):422-424. doi: 10.1111/imm.13831. Epub 2024 Jul 23. Immunology. 2024. PMID: 39039976 No abstract available.
References
Method references
-
- Barker N et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). https://doi.org/nature06196 [pii] 10.1038/nature06196 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

