An atlas of epithelial cell states and plasticity in lung adenocarcinoma
- PMID: 38418883
- PMCID: PMC10954546
- DOI: 10.1038/s41586-024-07113-9
An atlas of epithelial cell states and plasticity in lung adenocarcinoma
Erratum in
-
Author Correction: An atlas of epithelial cell states and plasticity in lung adenocarcinoma.Nature. 2024 Apr;628(8006):E1. doi: 10.1038/s41586-024-07277-4. Nature. 2024. PMID: 38499683 Free PMC article. No abstract available.
Abstract
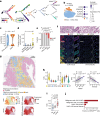
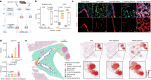
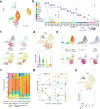
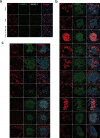
Understanding the cellular processes that underlie early lung adenocarcinoma (LUAD) development is needed to devise intervention strategies1. Here we studied 246,102 single epithelial cells from 16 early-stage LUADs and 47 matched normal lung samples. Epithelial cells comprised diverse normal and cancer cell states, and diversity among cancer cells was strongly linked to LUAD-specific oncogenic drivers. KRAS mutant cancer cells showed distinct transcriptional features, reduced differentiation and low levels of aneuploidy. Non-malignant areas surrounding human LUAD samples were enriched with alveolar intermediate cells that displayed elevated KRT8 expression (termed KRT8+ alveolar intermediate cells (KACs) here), reduced differentiation, increased plasticity and driver KRAS mutations. Expression profiles of KACs were enriched in lung precancer cells and in LUAD cells and signified poor survival. In mice exposed to tobacco carcinogen, KACs emerged before lung tumours and persisted for months after cessation of carcinogen exposure. Moreover, they acquired Kras mutations and conveyed sensitivity to targeted KRAS inhibition in KAC-enriched organoids derived from alveolar type 2 (AT2) cells. Last, lineage-labelling of AT2 cells or KRT8+ cells following carcinogen exposure showed that KACs are possible intermediates in AT2-to-tumour cell transformation. This study provides new insights into epithelial cell states at the root of LUAD development, and such states could harbour potential targets for prevention or intervention.
© 2024. The Author(s).
Conflict of interest statement
C.S.S. and A.E.S. are employees of Johnson & Johnson. H.K. reports research funding from Johnson & Johnson. M.V.N. receives research funding to institution from Mirati, Novartis, Checkmate, Alaunos/Ziopharm, AstraZeneca, Pfizer and Genentech, and consultant/advisory board fees from Mirati, Merck/MSD and Genentech. T.C. reports speaker fees/honoraria from The Society for Immunotherapy of Cancer, Bristol Myers Squibb, Roche, Medscape and PeerView; travel, food and beverage expenses from Dava Oncology and Bristol Myers Squibb; advisory role/consulting fees from MedImmune/AstraZeneca, Bristol Myers Squibb, EMD Serono, Merck & Co., Genentech, Arrowhead Pharmaceuticals and Regeneron; and institutional research funding from MedImmune/AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim and EMD Serono. S.J.M. reports funding from Arrowhead Pharma and Boehringer Ingelheim outside the scopes of submitted work. B.S. reports consulting and speaker fees from PeerView, AstraZeneca and Medscape, and institutional research funding from Bristol Myers Squibb. J.V.H. reports fees for advisory committees/consulting from AstraZeneca, EMD Serono, Boehringer-Ingelheim, Catalyst, Genentech, GlaxoSmithKline, Hengrui Therapeutics, Eli Lilly, Spectrum, Sanofi, Takeda, Mirati Therapeutics, BMS, BrightPath Biotherapeutics, Janssen Global Services, Nexus Health Systems, Pneuma Respiratory, Kairos Venture Investments, Roche, Leads Biolabs, RefleXion, Chugai Pharmaceuticals; research support from AstraZeneca, Bristol-Myers Squibb, Spectrum and Takeda, and royalties and licensing fees from Spectrum. I.I.W. reports grants and personal fees from Genentech/Roche, grants and personal fees from Bayer, grants and personal fees from Bristol-Myers Squibb, grants and personal fees from AstraZeneca, grants and personal fees from Pfizer, grants and personal fees from HTG Molecular, personal fees from Asuragen, grants and personal fees from Merck, grants and personal fees from GlaxoSmithKline, grants and personal fees from Guardant Health, personal fees from Flame, grants and personal fees from Novartis, grants and personal fees from Sanofi, personal fees from Daiichi Sankyo, grants and personal fees from Amgen, personal fees from Oncocyte, personal fees from MSD, personal fees from Platform Health, grants from Adaptive, grants from Adaptimmune, grants from EMD Serono, grants from Takeda, grants from Karus, grants from Johnson & Johnson, grants from 4D, from Iovance and from Akoya, outside the submitted work. All other authors declare no competing interests.
Figures













References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

