Cleaving DNA with DNA: Cooperative Tuning of Structure and Reactivity Driven by Copper Ions
- PMID: 38419268
- PMCID: PMC11040348
- DOI: 10.1002/advs.202306710
Cleaving DNA with DNA: Cooperative Tuning of Structure and Reactivity Driven by Copper Ions
Abstract
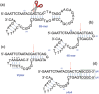
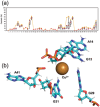
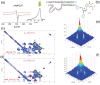
A copper-dependent self-cleaving DNA (DNAzyme or deoyxyribozyme) previously isolated by in vitro selection has been analyzed by a combination of Molecular Dynamics (MD) simulations and advanced Electron Paramagnetic Resonance (Electron Spin Resonance) EPR/ESR spectroscopy, providing insights on the structural and mechanistic features of the cleavage reaction. The modeled 46-nucleotide deoxyribozyme in MD simulations forms duplex and triplex sub-structures that flank a highly conserved catalytic core. The DNA self-cleaving construct can also form a bimolecular complex that has a distinct substrate and enzyme domains. The highly dynamic structure combined with an oxidative site-specific cleavage of the substrate are two key-aspects to elucidate. By combining EPR/ESR spectroscopy with selectively isotopically labeled nucleotides it has been possible to overcome the major drawback related to the "metal-soup" scenario, also known as "super-stoichiometric" ratios of cofactors versus substrate, conventionally required for the DNA cleavage reaction within those nucleic acids-based enzymes. The focus on the endogenous paramagnetic center (Cu2+) here described paves the way for analysis on mixtures where several different cofactors are involved. Furthermore, the insertion of cleavage reaction within more complex architectures is now a realistic perspective towards the applicability of EPR/ESR spectroscopic studies.
Keywords: DNAzymes; hyperfine spectroscopy; metal soup; multiple binding; radical path.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Characterization of a DNA-cleaving deoxyribozyme.Bioorg Med Chem. 2001 Oct;9(10):2589-600. doi: 10.1016/s0968-0896(01)00035-9. Bioorg Med Chem. 2001. PMID: 11557347
-
Sensitive dual DNAzymes-based sensors designed by grafting self-blocked G-quadruplex DNAzymes to the substrates of metal ion-triggered DNA/RNA-cleaving DNAzymes.Biosens Bioelectron. 2012 Oct-Dec;38(1):331-6. doi: 10.1016/j.bios.2012.06.011. Epub 2012 Jun 19. Biosens Bioelectron. 2012. PMID: 22784499
-
Nucleotide-Independent Copper(II)-Based Distance Measurements in DNA by Pulsed ESR Spectroscopy.Angew Chem Int Ed Engl. 2017 Feb 13;56(8):2115-2117. doi: 10.1002/anie.201611197. Epub 2017 Jan 16. Angew Chem Int Ed Engl. 2017. PMID: 28090713
-
Going the dHis-tance: Site-Directed Cu2+ Labeling of Proteins and Nucleic Acids.Acc Chem Res. 2021 Mar 16;54(6):1481-1491. doi: 10.1021/acs.accounts.0c00761. Epub 2021 Jan 21. Acc Chem Res. 2021. PMID: 33476119 Review.
-
Metal ion-dependent DNAzymes and their applications as biosensors.Met Ions Life Sci. 2012;10:217-48. doi: 10.1007/978-94-007-2172-2_8. Met Ions Life Sci. 2012. PMID: 22210341 Review.
Cited by
-
Quantity of Cu(II) ions in a copper pot by a DNAzyme-based fluorescent sensor.Food Chem (Oxf). 2025 Feb 18;10:100249. doi: 10.1016/j.fochms.2025.100249. eCollection 2025 Jun. Food Chem (Oxf). 2025. PMID: 40084088 Free PMC article.
References
-
- Sangeetha Gowda K. R., Blessy B. M., Sudhamani C. N., Bhoijya H. S., Biomedicine and Biotechnology 2014, 2, 1.
-
- Yuan R., Bickle T. A., Ebbers W., Brack C., Nature 1975, 256, 556. - PubMed
-
- Nagata S., Nagase H., Kawane K., Mukae N., Fukuyama H, Cell Death Differ. 2003, 10, 108. - PubMed
-
- Chen Y., Xu R., Chen J., Li X., He Q., Oncology Reports 2013, 30, 939. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
