miR-939-3p induces sarcoma proliferation and poor prognosis via suppressing BATF2
- PMID: 38420020
- PMCID: PMC10899471
- DOI: 10.3389/fonc.2024.1346531
miR-939-3p induces sarcoma proliferation and poor prognosis via suppressing BATF2
Abstract
Background: Sarcoma is a rare and aggressive malignancy with poor prognosis, in which oncogene activation and tumor suppressor inactivation are involved. Accumulated studies suggested basic leucine zipper transcription factor ATF-like 2 (BATF2) as a candidate tumor suppressor, but its specific role and mechanism in sarcoma remain unclear.
Methods: The expression levels of BATF2 and miR-939-3p were evaluated by using human sarcoma samples, cell lines and xenograft mouse models. Bioinformatics analysis, qPCR, Western blot, cell proliferation assay, overexpression plasmid construction, point mutation and dual luciferase reporter assay were utilized to investigate the role and mechanism of miR-939-3p in sarcoma.
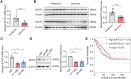
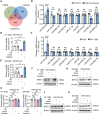
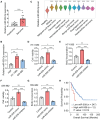
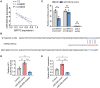
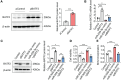
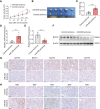
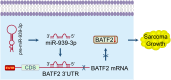
Results: In this study, we demonstrated that the expression of BATF2 was downregulated in human sarcoma tissues and cell lines. The downregulation of BATF2 was negatively associated with the prognosis of sarcoma patients. Subsequent bioinformatic prediction and experimental validations showed that BATF2 expression was reduced by microRNA (miR)-939-3p mimic and increased by miR-939-3p inhibitor. Additionally, miR-939-3p was upregulated in sarcoma tissues and cells, correlating with a poor prognosis of sarcoma patients. Moreover, miR-939-3p overexpression suppressed sarcoma cell proliferation, which was significantly attenuated by the restoration of BATF2, while siRNA-mediated knockdown of BATF2 aggravated the miR-939-3p-induced promotion of sarcoma cell proliferation. Further computational algorithms and dual-luciferase reporter assays demonstrated that miR-939-3p repressed BATF2 expression via directly binding to its 3' untranslated region (3' UTR).
Conclusion: Collectively, these findings identified miR-939-3p as a novel regulator of BATF2, as well as a prognostic biomarker in sarcoma, and revealed that suppressing miR-939-3p or inducing BATF2 expression may serve as a promising therapeutic strategy against sarcoma.
Keywords: BATF2; miR-939-3p; prognosis; sarcoma; therapeutic target.
Copyright © 2024 Xu, Huang, Lei and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Circ_0005927 Inhibits the Progression of Colorectal Cancer by Regulating miR-942-5p/BATF2 Axis.Cancer Manag Res. 2021 Mar 11;13:2295-2306. doi: 10.2147/CMAR.S281377. eCollection 2021. Cancer Manag Res. 2021. PMID: 33732022 Free PMC article.
-
Nuclear export of BATF2 enhances colorectal cancer proliferation through binding to CRM1.Clin Transl Med. 2023 May;13(5):e1260. doi: 10.1002/ctm2.1260. Clin Transl Med. 2023. PMID: 37151195 Free PMC article.
-
Bone marrow mesenchymal stem cells-derived exosomes suppress miRNA-5189-3p to increase fibroblast-like synoviocyte apoptosis via the BATF2/JAK2/STAT3 signaling pathway.Bioengineered. 2022 Mar;13(3):6767-6780. doi: 10.1080/21655979.2022.2045844. Bioengineered. 2022. PMID: 35246006 Free PMC article.
-
Down-regulation of EVA1A by miR-103a-3p promotes hepatocellular carcinoma cells proliferation and migration.Cell Mol Biol Lett. 2022 Oct 22;27(1):93. doi: 10.1186/s11658-022-00388-8. Cell Mol Biol Lett. 2022. PMID: 36273122 Free PMC article.
-
Role of the basic leucine zipper transcription factor BATF2 in modulating immune responses and inflammation in health and disease.J Leukoc Biol. 2025 Mar 14;117(3):qiae245. doi: 10.1093/jleuko/qiae245. J Leukoc Biol. 2025. PMID: 39504573 Review.
Cited by
-
Identification of novel diagnostic and prognostic microRNAs in sarcoma on TCGA dataset: bioinformatics and machine learning approach.Sci Rep. 2025 Mar 4;15(1):7521. doi: 10.1038/s41598-025-91007-x. Sci Rep. 2025. PMID: 40032929 Free PMC article.
References
LinkOut - more resources
Full Text Sources

