Chicken γδ T cells proliferate upon IL-2 and IL-12 treatment and show a restricted receptor repertoire in cell culture
- PMID: 38420118
- PMCID: PMC10900522
- DOI: 10.3389/fimmu.2024.1325024
Chicken γδ T cells proliferate upon IL-2 and IL-12 treatment and show a restricted receptor repertoire in cell culture
Abstract
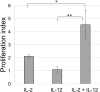
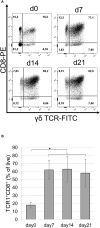
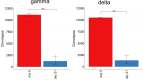
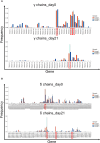
In chickens, γδ T cells represent a large fraction of peripheral T cells; however, their function remains largely unknown. Here, we describe the selective in vitro expansion of γδ T cells from total splenocytes by stimulation with the cytokines IL-2 and IL-12. Under these conditions, γδ T cells proliferated preferentially and reached frequencies of >95% within three weeks. Although IL-2 alone also triggered proliferation, an increased proliferation rate was observed in combination with IL-12. Most of the expanded cells were γδ TCR and CD8 double-positive. Splenocytes sorted into TCR1+CD8+, TCR1highCD8-, and TCR1lowCD8- subsets proliferated well upon dual stimulation with IL-2/IL-12, indicating that none of the three γδ T cell subsets require bystander activation for proliferation. TCR1+CD8+ cells maintained CD8 surface expression during stimulation, whereas CD8- subpopulations showed varied levels of CD8 upregulation, with the highest upregulation observed in the TCR1high subset. Changes in the γδ T-cell receptor repertoire during cell culture from day 0 to day 21 were analyzed by next-generation sequencing of the γδ variable regions. Overall, long-term culture led to a restricted γ and δ chain repertoire, characterized by a reduced number of unique variable region clonotypes, and specific V genes were enriched at day 21. On day 0, the δ chain repertoire was highly diverse, and the predominant clonotypes differed between animals, while the most frequent γ-chain clonotypes were shared between animals. However, on day 21, the most frequent clonotypes in both the γ and δ chain repertoires were different between animals, indicating that selective expansion of dominant clonotypes during stimulation seems to be an individual outcome. In conclusion, IL-2 and IL-12 were sufficient to stimulate the in vitro outgrowth of γδ T cells. Analyses of the TCR repertoire indicate that the culture leads to an expansion of individual T cell clones, which may reflect previous in vivo activation. This system will be instrumental in studying γδ T cell function.
Keywords: IL-12; IL-2; TCR repertoire analysis; chicken γδ TCR; in vitro culture; γδ T cell subsets.
Copyright © 2024 Linti, Göbel and Früh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

