Computational pathology: A survey review and the way forward
- PMID: 38420608
- PMCID: PMC10900832
- DOI: 10.1016/j.jpi.2023.100357
Computational pathology: A survey review and the way forward
Abstract
Computational Pathology (CPath) is an interdisciplinary science that augments developments of computational approaches to analyze and model medical histopathology images. The main objective for CPath is to develop infrastructure and workflows of digital diagnostics as an assistive CAD system for clinical pathology, facilitating transformational changes in the diagnosis and treatment of cancer that are mainly address by CPath tools. With evergrowing developments in deep learning and computer vision algorithms, and the ease of the data flow from digital pathology, currently CPath is witnessing a paradigm shift. Despite the sheer volume of engineering and scientific works being introduced for cancer image analysis, there is still a considerable gap of adopting and integrating these algorithms in clinical practice. This raises a significant question regarding the direction and trends that are undertaken in CPath. In this article we provide a comprehensive review of more than 800 papers to address the challenges faced in problem design all-the-way to the application and implementation viewpoints. We have catalogued each paper into a model-card by examining the key works and challenges faced to layout the current landscape in CPath. We hope this helps the community to locate relevant works and facilitate understanding of the field's future directions. In a nutshell, we oversee the CPath developments in cycle of stages which are required to be cohesively linked together to address the challenges associated with such multidisciplinary science. We overview this cycle from different perspectives of data-centric, model-centric, and application-centric problems. We finally sketch remaining challenges and provide directions for future technical developments and clinical integration of CPath. For updated information on this survey review paper and accessing to the original model cards repository, please refer to GitHub. Updated version of this draft can also be found from arXiv.
Keywords: Clinical pathology; Computer aided diagnosis (CAD); Deep learning; Digital pathology; Survey; Whole slide image (WSI).
© 2024 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
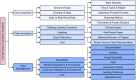
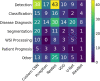
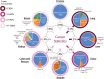
Figures





References
-
- FDA News Release Fda allows marketing of first whole slide imaging system for digital pathology. 2017. https://www.fda.gov/news-events/press-announcements/fda-allows-marketing...
-
- Evans Andrew J., Bauer Thomas W., Bui Marilyn M., et al. Us food and drug administration approval of whole slide imaging for primary diagnosis: a key milestone is reached and new questions are raised. Arch Pathol Lab Med. 2018;142(11):1383–1387. - PubMed
-
- Araújo Anna Luíza Damaceno, Arboleda Lady Paola Aristizábal, Palmier Natalia Rangel, et al. The performance of digital microscopy for primary diagnosis in human pathology: a systematic review. Virchows Arch. 2019;474(3):269–287. - PubMed
-
- Williams Bethany Jill, Hanby Andrew, Millican-Slater Rebecca, Nijhawan Anju, Verghese Eldo, Treanor Darren. Digital pathology for the primary diagnosis of breast histopathological specimens: an innovative validation and concordance study on digital pathology validation and training. Histopathology. 2018;72(4):662–671. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

