Type I Interferon Autoantibodies Correlate With Cellular Immune Alterations in Severe COVID-19
- PMID: 38421006
- PMCID: PMC11326830
- DOI: 10.1093/infdis/jiae036
Type I Interferon Autoantibodies Correlate With Cellular Immune Alterations in Severe COVID-19
Abstract
Background: Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can lead to severe disease with increased morbidity and mortality among certain risk groups. The presence of autoantibodies against type I interferons (aIFN-Abs) is one mechanism that contributes to severe coronavirus disease 2019 (COVID-19).
Methods: This study aimed to investigate the presence of aIFN-Abs in relation to the soluble proteome, circulating immune cell numbers, and cellular phenotypes, as well as development of adaptive immunity.
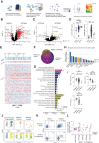
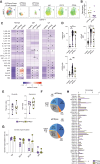
Results: aIFN-Abs were more prevalent in critical compared to severe COVID-19 but largely absent in the other viral and bacterial infections studied here. The antibody and T-cell response to SARS-CoV-2 remained largely unaffected by the presence aIFN-Abs. Similarly, the inflammatory response in COVID-19 was comparable in individuals with and without aIFN-Abs. Instead, presence of aIFN-Abs had an impact on cellular immune system composition and skewing of cellular immune pathways.
Conclusions: Our data suggest that aIFN-Abs do not significantly influence development of adaptive immunity but covary with alterations in immune cell numbers.
Keywords: COVID-19; autoantibodies; immunity; interferon.
© The Author(s) 2024. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest . S. A. has received honoraria for lectures and educational events from Gilead, AbbVie, MSD, and Biogen; and reports grants from Gilead and AbbVie. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures

References
-
- Galani I-E, Rovina N, Lampropoulou V, et al. . Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol 2021; 22:32–40. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

