Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus
- PMID: 38421610
- PMCID: PMC11040149
- DOI: 10.1093/nar/gkae149
Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus
Abstract
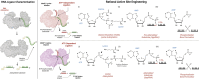
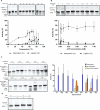
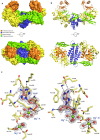
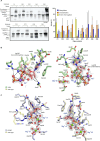
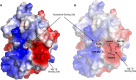
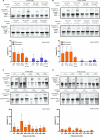
RNA ligases are important enzymes in molecular biology and are highly useful for the manipulation and analysis of nucleic acids, including adapter ligation in next-generation sequencing of microRNAs. Thermophilic RNA ligases belonging to the RNA ligase 3 family are gaining attention for their use in molecular biology, for example a thermophilic RNA ligase from Methanobacterium thermoautotrophicum is commercially available for the adenylation of nucleic acids. Here we extensively characterise a newly identified RNA ligase from the thermophilic archaeon Palaeococcus pacificus (PpaRnl). PpaRnl exhibited significant substrate adenylation activity but low ligation activity across a range of oligonucleotide substrates. Mutation of Lys92 in motif I to alanine, resulted in an enzyme that lacked adenylation activity, but demonstrated improved ligation activity with pre-adenylated substrates (ATP-independent ligation). Subsequent structural characterisation revealed that in this mutant enzyme Lys238 was found in two alternate positions for coordination of the phosphate tail of ATP. In contrast mutation of Lys238 in motif V to glycine via structure-guided engineering enhanced ATP-dependent ligation activity via an arginine residue compensating for the absence of Lys238. Ligation activity for both mutations was higher than the wild-type, with activity observed across a range of oligonucleotide substrates with varying sequence and secondary structure.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Structure-function analysis of Methanobacterium thermoautotrophicum RNA ligase - engineering a thermostable ATP independent enzyme.BMC Mol Biol. 2012 Jul 18;13:24. doi: 10.1186/1471-2199-13-24. BMC Mol Biol. 2012. PMID: 22809063 Free PMC article.
-
Kinetic and structural insights into the requirement of fungal tRNA ligase for a 2'-phosphate end.RNA. 2024 Sep 16;30(10):1306-1314. doi: 10.1261/rna.080120.124. RNA. 2024. PMID: 39013577
-
The rad50 signature motif: essential to ATP binding and biological function.J Mol Biol. 2004 Jan 23;335(4):937-51. doi: 10.1016/j.jmb.2003.11.026. J Mol Biol. 2004. PMID: 14698290
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
Cited by
-
An RNA ligase partner for the prokaryotic protein-only RNase P: insights into the functional diversity of RNase P from genome mining.mBio. 2025 Jun 11;16(6):e0044925. doi: 10.1128/mbio.00449-25. Epub 2025 Apr 29. mBio. 2025. PMID: 40298408 Free PMC article.
References
-
- Pascal J.M. DNA and RNA ligases: structural variations and shared mechanisms. Curr. Opin. Struct. Biol. 2008; 18:96–105. - PubMed
-
- Nichols N.M., Tabor S., McReynolds L.A. RNA Ligases. Curr. Protoc. Mol. Biol. 2008; 84:3.15.1–3.15.4. - PubMed
-
- Ho C.K., Wang L.K., Lima C.D., Shuman S. Structure and mechanism of RNA ligase. Structure. 2004; 12:327–339. - PubMed
-
- Wood Z.A., Sabatini R.S., Hajduk S.L. RNA ligase: picking up the pieces. Mol. Cell. 2004; 13:455–456. - PubMed
-
- Shuman S., Lima C.D. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004; 14:757–764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

