The repertoire and structure of adhesion GPCR transcript variants assembled from publicly available deep-sequenced human samples
- PMID: 38421639
- PMCID: PMC11039983
- DOI: 10.1093/nar/gkae145
The repertoire and structure of adhesion GPCR transcript variants assembled from publicly available deep-sequenced human samples
Abstract
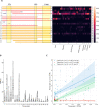
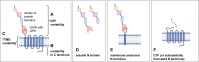
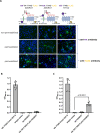
Alternative splicing and multiple transcription start and termination sites can produce a diverse repertoire of mRNA transcript variants from a given gene. While the full picture of the human transcriptome is still incomplete, publicly available RNA datasets have enabled the assembly of transcripts. Using publicly available deep sequencing data from 927 human samples across 48 tissues, we quantified known and new transcript variants, provide an interactive, browser-based application Splice-O-Mat and demonstrate its relevance using adhesion G protein-coupled receptors (aGPCRs) as an example. On average, 24 different transcript variants were detected for each of the 33 human aGPCR genes, and several dominant transcript variants were not yet annotated. Variable transcription starts and complex exon-intron structures encode a flexible protein domain architecture of the N- and C termini and the seven-transmembrane helix domain (7TMD). Notably, we discovered the first GPCR (ADGRG7/GPR128) with eight transmembrane helices. Both the N- and C terminus of this aGPCR were intracellularly oriented, anchoring the N terminus in the plasma membrane. Moreover, the assessment of tissue-specific transcript variants, also for other gene classes, in our application may change the evaluation of disease-causing mutations, as their position in different transcript variants may explain tissue-specific phenotypes.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- McMillan D.R., Kayes-Wandover K.M., Richardson J.A., White P.C. Very large G protein-coupled receptor-1, the largest known cell surface protein, is highly expressed in the developing central nervous system. J. Biol. Chem. 2002; 277:785–792. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

