SynGAP regulates synaptic plasticity and cognition independently of its catalytic activity
- PMID: 38422154
- PMCID: PMC11188940
- DOI: 10.1126/science.adk1291
SynGAP regulates synaptic plasticity and cognition independently of its catalytic activity
Abstract
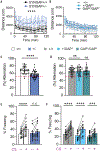
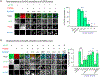
SynGAP is an abundant synaptic GTPase-activating protein (GAP) critical for synaptic plasticity, learning, memory, and cognition. Mutations in SYNGAP1 in humans result in intellectual disability, autistic-like behaviors, and epilepsy. Heterozygous Syngap1-knockout mice display deficits in synaptic plasticity, learning, and memory and exhibit seizures. It is unclear whether SynGAP imparts structural properties at synapses independently of its GAP activity. Here, we report that inactivating mutations within the GAP domain do not inhibit synaptic plasticity or cause behavioral deficits. Instead, SynGAP modulates synaptic strength by physically competing with the AMPA-receptor-TARP excitatory receptor complex in the formation of molecular condensates with synaptic scaffolding proteins. These results have major implications for developing therapeutic treatments for SYNGAP1-related neurodevelopmental disorders.
Figures






Comment in
-
Shifting rules in a brain disorder.Science. 2024 Mar;383(6686):950-951. doi: 10.1126/science.adn8707. Epub 2024 Feb 29. Science. 2024. PMID: 38422158
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

