Vocal learning-associated convergent evolution in mammalian proteins and regulatory elements
- PMID: 38422184
- PMCID: PMC11313673
- DOI: 10.1126/science.abn3263
Vocal learning-associated convergent evolution in mammalian proteins and regulatory elements
Abstract
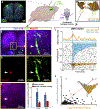
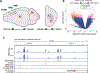
Vocal production learning ("vocal learning") is a convergently evolved trait in vertebrates. To identify brain genomic elements associated with mammalian vocal learning, we integrated genomic, anatomical, and neurophysiological data from the Egyptian fruit bat (Rousettus aegyptiacus) with analyses of the genomes of 215 placental mammals. First, we identified a set of proteins evolving more slowly in vocal learners. Then, we discovered a vocal motor cortical region in the Egyptian fruit bat, an emergent vocal learner, and leveraged that knowledge to identify active cis-regulatory elements in the motor cortex of vocal learners. Machine learning methods applied to motor cortex open chromatin revealed 50 enhancers robustly associated with vocal learning whose activity tended to be lower in vocal learners. Our research implicates convergent losses of motor cortex regulatory elements in mammalian vocal learning evolution.
Conflict of interest statement
Figures


References
-
- Doupe AJ, Kuhl PK, Birdsong and human speech: common themes and mechanisms. Annu. Rev. Neurosci. 22, 567–631 (1999). - PubMed
-
- Pfenning AR, Hara E, Whitney O, Rivas MV, Wang R, Roulhac PL, Howard JT, Wirthlin M, Lovell PV, Ganapathy G, Mouncastle J, Moseley MA, Thompson JW, Soderblom EJ, Iriki A, Kato M, Gilbert MTP, Zhang G, Bakken T, Bongaarts A, Bernard A, Lein E, Mello CV, Hartemink AJ, Jarvis ED, Convergent transcriptional specializations in the brains of humans and song-learning birds. Science 346, 1256846 (2014). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

