Inflammation and bacteriophages affect DNA inversion states and functionality of the gut microbiota
- PMID: 38423015
- PMCID: PMC10939037
- DOI: 10.1016/j.chom.2024.02.003
Inflammation and bacteriophages affect DNA inversion states and functionality of the gut microbiota
Abstract
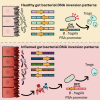
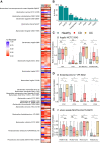
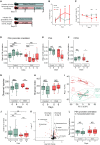
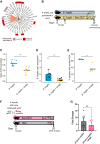
Reversible genomic DNA inversions control the expression of numerous gut bacterial molecules, but how this impacts disease remains uncertain. By analyzing metagenomic samples from inflammatory bowel disease (IBD) cohorts, we identified multiple invertible regions where a particular orientation correlated with disease. These include the promoter of polysaccharide A (PSA) of Bacteroides fragilis, which induces regulatory T cells (Tregs) and ameliorates experimental colitis. The PSA promoter was mostly oriented "OFF" in IBD patients, which correlated with increased B. fragilis-associated bacteriophages. Similarly, in mice colonized with a healthy human microbiota and B. fragilis, induction of colitis caused a decline of PSA in the "ON" orientation that reversed as inflammation resolved. Monocolonization of mice with B. fragilis revealed that bacteriophage infection increased the frequency of PSA in the "OFF" orientation, causing reduced PSA expression and decreased Treg cells. Altogether, we reveal dynamic bacterial phase variations driven by bacteriophages and host inflammation, signifying bacterial functional plasticity during disease.
Keywords: Bacteroides; Crohn’s disease; DNA inversions; bacteriophages; functional plasticity; gut microbiome; immunomodulation; inflammatory bowel diseases; phase variation; ulcerative colitis.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Moxon R., Bayliss C., Hood D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 2006;40:307–333. - PubMed
-
- Phillips Z.N., Tram G., Seib K.L., Atack J.M. Phase-variable bacterial loci: how bacteria gamble to maximise fitness in changing environments. Biochem. Soc. Trans. 2019;47:1131–1141. - PubMed
-
- Krinos C.M., Coyne M.J., Weinacht K.G., Tzianabos A.O., Kasper D.L., Comstock L.E. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414:555–558. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

