MYC induces CDK4/6 inhibitors resistance by promoting pRB1 degradation
- PMID: 38424044
- PMCID: PMC10904810
- DOI: 10.1038/s41467-024-45796-w
MYC induces CDK4/6 inhibitors resistance by promoting pRB1 degradation
Abstract
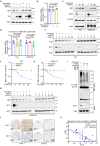
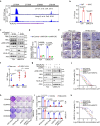
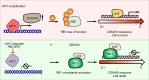
CDK4/6 inhibitors (CDK4/6i) show anticancer activity in certain human malignancies, such as breast cancer. However, their application to other tumor types and intrinsic resistance mechanisms are still unclear. Here, we demonstrate that MYC amplification confers resistance to CDK4/6i in bladder, prostate and breast cancer cells. Mechanistically, MYC binds to the promoter of the E3 ubiquitin ligase KLHL42 and enhances its transcription, leading to RB1 deficiency by inducing both phosphorylated and total pRB1 ubiquitination and degradation. We identify a compound that degrades MYC, A80.2HCl, which induces MYC degradation at nanomolar concentrations, restores pRB1 protein levels and re-establish sensitivity of MYC high-expressing cancer cells to CDK4/6i. The combination of CDK4/6i and A80.2HCl result in marked regression in tumor growth in vivo. Altogether, these results reveal the molecular mechanisms underlying MYC-induced resistance to CDK4/6i and suggest the utilization of the MYC degrading molecule A80.2HCl to potentiate the therapeutic efficacy of CDK4/6i.
© 2024. The Author(s).
Conflict of interest statement
W.W. is a co-founder and consultant for the ReKindle Therapeutics. Z.R. is the VP of Kintor Pharmaceutical, Inc. Other authors declare no competing interests.
Figures




References
-
- Patnaik A, et al. Efficacy and safety of Abemaciclib, an Inhibitor of CDK4 and CDK6, for patients with breast cancer, non-small cell lung cancer, and other solid tumors. Cancer Discov. 2016;6:740–753. doi: 10.1158/2159-8290.CD-16-0095. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 81925028/National Natural Science Foundation of China (National Science Foundation of China)
- 82230097/National Natural Science Foundation of China (National Science Foundation of China)
- 82173037/National Natural Science Foundation of China (National Science Foundation of China)
- 82273106/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

