Exploring the gut DNA virome in fecal immunochemical test stool samples reveals associations with lifestyle in a large population-based study
- PMID: 38424056
- PMCID: PMC10904388
- DOI: 10.1038/s41467-024-46033-0
Exploring the gut DNA virome in fecal immunochemical test stool samples reveals associations with lifestyle in a large population-based study
Abstract
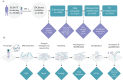
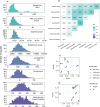
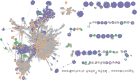
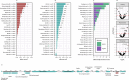
Stool samples for fecal immunochemical tests (FIT) are collected in large numbers worldwide as part of colorectal cancer screening programs. Employing FIT samples from 1034 CRCbiome participants, recruited from a Norwegian colorectal cancer screening study, we identify, annotate and characterize more than 18000 DNA viruses, using shotgun metagenome sequencing. Only six percent of them are assigned to a known taxonomic family, with Microviridae being the most prevalent viral family. Linking individual profiles to comprehensive lifestyle and demographic data shows 17/25 of the variables to be associated with the gut virome. Physical activity, smoking, and dietary fiber consumption exhibit strong and consistent associations with both diversity and relative abundance of individual viruses, as well as with enrichment for auxiliary metabolic genes. We demonstrate the suitability of FIT samples for virome analysis, opening an opportunity for large-scale studies of this enigmatic part of the gut microbiome. The diverse viral populations and their connections to the individual lifestyle uncovered herein paves the way for further exploration of the role of the gut virome in health and disease.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Meta-analysis of fecal viromes demonstrates high diagnostic potential of the gut viral signatures for colorectal cancer and adenoma risk assessment.J Adv Res. 2023 Jul;49:103-114. doi: 10.1016/j.jare.2022.09.012. Epub 2022 Oct 2. J Adv Res. 2023. PMID: 36198381 Free PMC article.
-
Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes.Gastroenterology. 2018 Aug;155(2):529-541.e5. doi: 10.1053/j.gastro.2018.04.018. Epub 2018 Apr 22. Gastroenterology. 2018. PMID: 29689266
-
The Chinese gut virus catalogue reveals gut virome diversity and disease-related viral signatures.Genome Med. 2025 Mar 26;17(1):30. doi: 10.1186/s13073-025-01460-6. Genome Med. 2025. PMID: 40140988 Free PMC article.
-
The gut virome in inflammatory bowel diseases.Curr Opin Virol. 2021 Dec;51:190-198. doi: 10.1016/j.coviro.2021.10.005. Epub 2021 Nov 8. Curr Opin Virol. 2021. PMID: 34763180 Review.
-
The Human Gut Virome and Its Relationship with Nontransmissible Chronic Diseases.Nutrients. 2023 Feb 15;15(4):977. doi: 10.3390/nu15040977. Nutrients. 2023. PMID: 36839335 Free PMC article. Review.
Cited by
-
Fecal microbiomes from screening sampling tubes are stable despite varying sampling and storage conditions.Sci Rep. 2025 Jul 24;15(1):26951. doi: 10.1038/s41598-025-12506-5. Sci Rep. 2025. PMID: 40707563 Free PMC article.
-
Species-level verification of Phascolarctobacterium association with colorectal cancer.mSystems. 2024 Oct 22;9(10):e0073424. doi: 10.1128/msystems.00734-24. Epub 2024 Sep 17. mSystems. 2024. PMID: 39287376 Free PMC article.
-
The Aggregated Gut Viral Catalogue (AVrC): A unified resource for exploring the viral diversity of the human gut.PLoS Comput Biol. 2025 May 2;21(5):e1012268. doi: 10.1371/journal.pcbi.1012268. eCollection 2025 May. PLoS Comput Biol. 2025. PMID: 40315414 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

