Deregulated protein homeostasis constrains fetal hematopoietic stem cell pool expansion in Fanconi anemia
- PMID: 38424108
- PMCID: PMC10904799
- DOI: 10.1038/s41467-024-46159-1
Deregulated protein homeostasis constrains fetal hematopoietic stem cell pool expansion in Fanconi anemia
Abstract
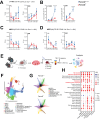
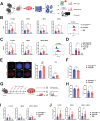
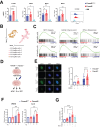
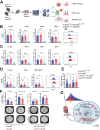
Demand-adjusted and cell type specific rates of protein synthesis represent an important safeguard for fate and function of long-term hematopoietic stem cells. Here, we identify increased protein synthesis rates in the fetal hematopoietic stem cell pool at the onset of hematopoietic failure in Fanconi Anemia, a prototypical DNA repair disorder that manifests with bone marrow failure. Mechanistically, the accumulation of misfolded proteins in Fancd2-/- fetal liver hematopoietic stem cells converges on endoplasmic reticulum stress, which in turn constrains midgestational expansion. Restoration of protein folding by the chemical chaperone tauroursodeoxycholic acid, a hydrophilic bile salt, prevents accumulation of unfolded proteins and rescues Fancd2-/- fetal liver long-term hematopoietic stem cell numbers. We find that proteostasis deregulation itself is driven by excess sterile inflammatory activity in hematopoietic and stromal cells within the fetal liver, and dampened Type I interferon signaling similarly restores fetal Fancd2-/- long-term hematopoietic stem cells to wild type-equivalent numbers. Our study reveals the origin and pathophysiological trigger that gives rise to Fanconi anemia hematopoietic stem cell pool deficits. More broadly, we show that fetal protein homeostasis serves as a physiological rheostat for hematopoietic stem cell fate and function.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Fancd2 and p21 function independently in maintaining the size of hematopoietic stem and progenitor cell pool in mice.Stem Cell Res. 2013 Sep;11(2):687-92. doi: 10.1016/j.scr.2013.04.010. Epub 2013 Apr 30. Stem Cell Res. 2013. PMID: 23721813 Free PMC article.
-
Radiologic differences between bone marrow stromal and hematopoietic progenitor cell lines from Fanconi Anemia (Fancd2(-/-)) mice.Radiat Res. 2014 Jan;181(1):76-89. doi: 10.1667/RR13405.1. Epub 2014 Jan 7. Radiat Res. 2014. PMID: 24397476 Free PMC article.
-
Endogenous DNA Damage Leads to p53-Independent Deficits in Replicative Fitness in Fetal Murine Fancd2-/- Hematopoietic Stem and Progenitor Cells.Stem Cell Reports. 2016 Nov 8;7(5):840-853. doi: 10.1016/j.stemcr.2016.09.005. Epub 2016 Oct 6. Stem Cell Reports. 2016. PMID: 27720904 Free PMC article.
-
The Fanconi anemia ID2 complex: dueling saxes at the crossroads.Cell Cycle. 2014;13(19):2999-3015. doi: 10.4161/15384101.2014.956475. Cell Cycle. 2014. PMID: 25486561 Free PMC article. Review.
-
Gene therapy for fanconi anemia.Curr Hematol Rep. 2003 Jul;2(4):335-40. Curr Hematol Rep. 2003. PMID: 12901331 Review.
Cited by
-
Inhibition of DEK restores hematopoietic stem cell function in Fanconi anemia.J Exp Med. 2025 Mar 3;222(3):e20241248. doi: 10.1084/jem.20241248. Epub 2025 Jan 21. J Exp Med. 2025. PMID: 39836085
-
The Fanconi Anemia Pathway Inhibits mTOR Signaling and Prevents Accelerated Translation in Head and Neck Cancer Cells.Cancers (Basel). 2025 Aug 6;17(15):2583. doi: 10.3390/cancers17152583. Cancers (Basel). 2025. PMID: 40805278 Free PMC article.
-
EMP1 safeguards hematopoietic stem cells by suppressing sphingolipid metabolism and alleviating endoplasmic reticulum stress.Nat Commun. 2025 Jul 7;16(1):6247. doi: 10.1038/s41467-025-61552-0. Nat Commun. 2025. PMID: 40624017 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

