CKAP5 stabilizes CENP-E at kinetochores by regulating microtubule-chromosome attachments
- PMID: 38424231
- PMCID: PMC11014917
- DOI: 10.1038/s44319-024-00106-9
CKAP5 stabilizes CENP-E at kinetochores by regulating microtubule-chromosome attachments
Abstract
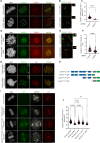
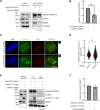
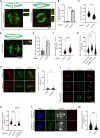
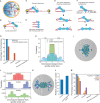
Stabilization of microtubule plus end-directed kinesin CENP-E at the metaphase kinetochores is important for chromosome alignment, but its mechanism remains unclear. Here, we show that CKAP5, a conserved microtubule plus tip protein, regulates CENP-E at kinetochores in human cells. Depletion of CKAP5 impairs CENP-E localization at kinetochores at the metaphase plate and results in increased kinetochore-microtubule stability and attachment errors. Erroneous attachments are also supported by computational modeling. Analysis of CKAP5 knockout cancer cells of multiple tissue origins shows that CKAP5 is preferentially essential in aneuploid, chromosomally unstable cells, and the sensitivity to CKAP5 depletion is correlated to that of CENP-E depletion. CKAP5 depletion leads to reduction in CENP-E-BubR1 interaction and the interaction is rescued by TOG4-TOG5 domain of CKAP5. The same domain can rescue CKAP5 depletion-induced CENP-E removal from the kinetochores. Interestingly, CKAP5 depletion facilitates recruitment of PP1 to the kinetochores and furthermore, a PP1 target site-specific CENP-E phospho-mimicking mutant gets stabilized at kinetochores in the CKAP5-depleted cells. Together, the results support a model in which CKAP5 controls mitotic chromosome attachment errors by stabilizing CENP-E at kinetochores and by regulating stability of the kinetochore-attached microtubules.
Keywords: CENP-E; CKAP5; Kinetochore; Microtubule; PP1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

