Progesterone receptor potentiates macropinocytosis through CDC42 in pancreatic ductal adenocarcinoma
- PMID: 38424455
- PMCID: PMC10904380
- DOI: 10.1038/s41389-024-00512-7
Progesterone receptor potentiates macropinocytosis through CDC42 in pancreatic ductal adenocarcinoma
Abstract
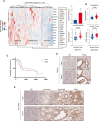
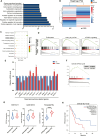
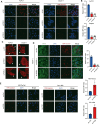
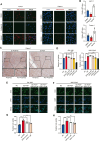
Endocrine receptors play an essential role in tumor metabolic reprogramming and represent a promising therapeutic avenue in pancreatic ductal adenocarcinoma (PDAC). PDAC is characterized by a nutrient-deprived microenvironment. To meet their ascendant energy demands, cancer cells can internalize extracellular proteins via macropinocytosis. However, the roles of endocrine receptors in macropinocytosis are not clear. In this study, we found that progesterone receptor (PGR), a steroid-responsive nuclear receptor, is highly expressed in PDAC tissues obtained from both patients and transgenic LSL-KrasG12D/+; LSL-Trp53R172H/+; PDX1-cre (KPC) mice. Moreover, PGR knockdown restrained PDAC cell survival and tumor growth both in vitro and in vivo. Genetic and pharmacological PGR inhibition resulted in a marked attenuation of macropinocytosis in PDAC cells and subcutaneous tumor models, indicating the involvement of this receptor in macropinocytosis regulation. Mechanistically, PGR upregulated CDC42, a critical regulator in macropinocytosis, through PGR-mediated transcriptional activation. These data deepen the understanding of how the endocrine system influences tumor progression via a non-classical pathway and provide a novel therapeutic option for patients with PDAC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
A low amino acid environment promotes cell macropinocytosis through the YY1-FGD6 axis in Ras-mutant pancreatic ductal adenocarcinoma.Oncogene. 2022 Feb;41(8):1203-1215. doi: 10.1038/s41388-021-02159-9. Epub 2022 Jan 27. Oncogene. 2022. PMID: 35082383
-
RAGE gene deletion inhibits the development and progression of ductal neoplasia and prolongs survival in a murine model of pancreatic cancer.J Gastrointest Surg. 2012 Jan;16(1):104-12; discussion 112. doi: 10.1007/s11605-011-1754-9. Epub 2011 Nov 4. J Gastrointest Surg. 2012. PMID: 22052106 Free PMC article.
-
Regulator of Chromosome Condensation (RCC1) a novel therapeutic target in pancreatic ductal adenocarcinoma drives tumor progression via the c-Myc-RCC1-Ran axis.bioRxiv [Preprint]. 2023 Dec 19:2023.12.18.572102. doi: 10.1101/2023.12.18.572102. bioRxiv. 2023. PMID: 38187605 Free PMC article. Preprint.
-
Dinaciclib prolongs survival in the LSL-KrasG12D/+ ; LSL-Trp53R172H/+ ; Pdx-1-Cre (KPC) transgenic murine models of pancreatic ductal adenocarcinoma.Am J Transl Res. 2020 Mar 15;12(3):1031-1043. eCollection 2020. Am J Transl Res. 2020. PMID: 32269732 Free PMC article.
-
Differences between KC and KPC pancreatic ductal adenocarcinoma mice models, in terms of their modeling biology and their clinical relevance.Pancreatology. 2020 Jan;20(1):79-88. doi: 10.1016/j.pan.2019.11.006. Epub 2019 Nov 18. Pancreatology. 2020. PMID: 31780287 Review.
Cited by
-
Sex Matters-Insights from Testing Drug Efficacy in an Animal Model of Pancreatic Cancer.Cancers (Basel). 2024 May 16;16(10):1901. doi: 10.3390/cancers16101901. Cancers (Basel). 2024. PMID: 38791980 Free PMC article.
References
-
- Kleeff, J, Korc, M, Apte, M, La Vecchia, C, Johnson, CD, Biankin, AV, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. 10.1038/nrdp.2016.22. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

