A combinatorial genetic strategy for exploring complex genotype-phenotype associations in cancer
- PMID: 38424461
- PMCID: PMC10937382
- DOI: 10.1038/s41588-024-01674-1
A combinatorial genetic strategy for exploring complex genotype-phenotype associations in cancer
Abstract
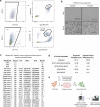
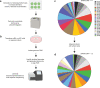
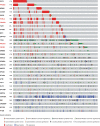
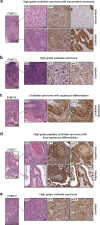
Available genetically defined cancer models are limited in genotypic and phenotypic complexity and underrepresent the heterogeneity of human cancer. Here, we describe a combinatorial genetic strategy applied to an organoid transformation assay to rapidly generate diverse, clinically relevant bladder and prostate cancer models. Importantly, the clonal architecture of the resultant tumors can be resolved using single-cell or spatially resolved next-generation sequencing to uncover polygenic drivers of cancer phenotypes.
© 2024. The Author(s).
Conflict of interest statement
J.K.L. served on the Speaker’s Bureau for Mission Bio. B.M.F. has consulting or advisory roles with QED Therapeutics, Boston Gene, Astrin Biosciences Merck, Immunomedics/Gilead, Guardant and Janssen and receives patent royalties from Immunomedics/Gilead and research support from Eli Lilly. All other authors declare no competing interests.
Figures




Update of
-
Combinatorial genetic strategy accelerates the discovery of cancer genotype-phenotype associations.bioRxiv [Preprint]. 2023 Apr 14:2023.04.12.536652. doi: 10.1101/2023.04.12.536652. bioRxiv. 2023. Update in: Nat Genet. 2024 Mar;56(3):371-376. doi: 10.1038/s41588-024-01674-1. PMID: 37609344 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

